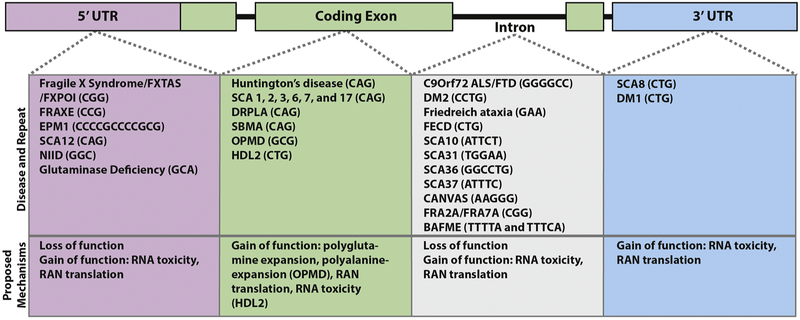
Figure 1: Nucleotide repeat expansion disorders.
Disorders are listed by their genomic location on an illustrated simplified gene. Each disorder is accompanied by the unstable repeat sequence that elicits disease. The prevailing disease mechanism is listed below each group to illustrate that genomic location of the expansion influences but does not completely determine the potential effects of any given repeat. Abbreviations: Fragile X-associated tremor/ataxia syndrome (FXTAS), Fragile X-primary ovarian insufficiency (FXPOI), progressive myoclonic epilepsy type 1/Unverricht-Lundborg disease (EPM1), spinocerebellar ataxia (SCA), neuronal intranuclear inclusion disease (NIID), dentatorubral-pallidoluysian atrophy (DRPLA), spinal-bulbar muscular atrophy (SBMA), oculopharyngeal muscular dystrophy (OPMD), Huntington disease-like 2 (HDL2), amyotrophic lateral sclerosis (ALS), frontotemporal dementia (FTD), myotonic dystrophy (DM), Fuchs endothelial corneal dystrophy (FECD), cerebellar ataxia, neuropathy and vestibular areflexia yndrome (CANVAS), benign adult familial myoclonic epilepsy (BAFME), and FRAXE, FRA2A and FRA7A refer to disease associated fragile sites in the genome that result from CGG repeat expansions.