Fig. 1.
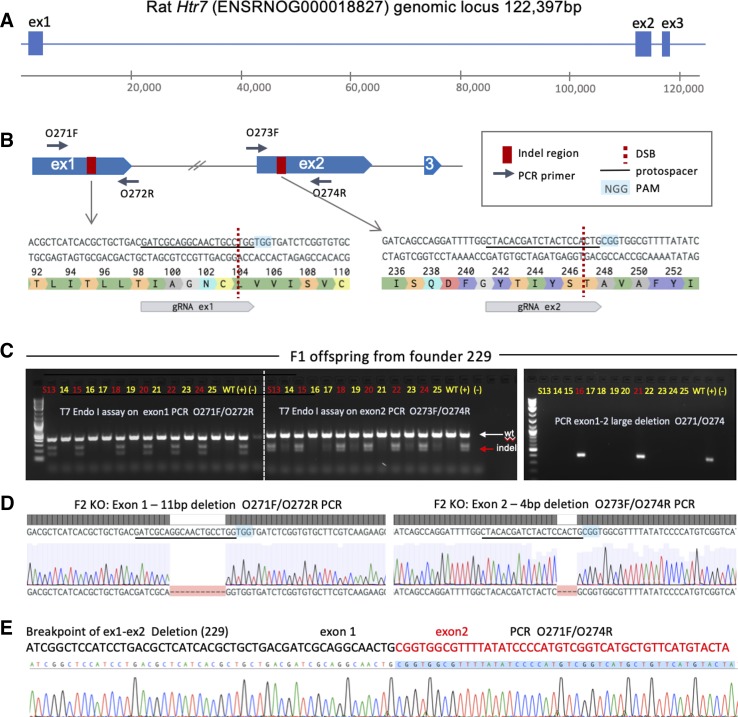
Targeting of the Htr7 locus. A: the rat Htr7 genomic locus (exon size not to scale), exon 1 and exon 2 were targeted by CRISPR-Cas9. B: location and exact sequence of gRNA targets within exon 1 and exon 2, primer design to detect indels, as well as large deletion are indicated. Location of DSBs and PAMs are indicated. C: raw gel electrophoresis images of T7 Endonuclease I (T7 Endo I) digestion analysis of F1 offspring from founder 229 carrying ex1 and ex2 indels on one allele, and a large deletion between ex1 and ex2 DSB sites on the second allele. D: individual gene edits from founder 229 and its offspring were characterized by Sanger sequencing of indicated PCR products: an 11 bp deletion in exon 1 and a 4 bp deletion in exon 2 were identified in F2 homozygous knockout (KO) animals. Location of deletions with respect to gRNA protospacer and PAM are indicated. E: exact sequence and breakpoint of a large deletion between exon 1 and exon 2 DSB sites are indicated. DSB, double-strand DNA break; PAM, protospacer adjacent motif.