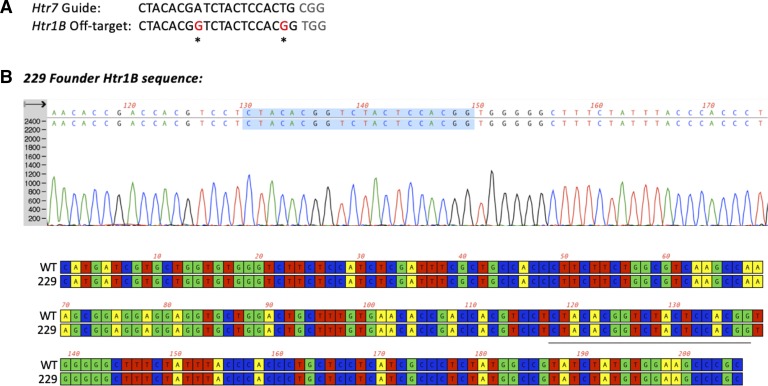
Fig. 2.
Off-target analysis. A: gRNA targeting exon 2 of the Htr7 locus also recognizes a target in the Htr1B locus with two mismatches (CFD off-target score: 0.183, MIT off-target score: 2.93, position: chr9:81631922–81631944). To rule out that an off-target hit occurred in the Htr1B locus during Htr7 editing, we performed PCR and Sanger sequencing of the amplicon surrounding the potential off-target site, with primers O409 and O410 (Table 1). Genomic DNA from founder 229 was used for PCR amplification. B: Sanger sequencing and alignment between wild-type (WT) and 5-HT7 KO 229 founder revealed no changes at the predicted cut site or nearby bases, indicating that no off-target changes at the Htr1B locus have occurred. Protospacer of off-target gRNA is underlined or highlighted in sequence and alignment.