Significance
The question of how transcription factors establish their preference for in vivo target loci on a genome-wide level has remained elusive, particularly for paralogous factors, which often prefer highly similar DNA motifs. The conceptual in vivo specificity models put forward, including cooperativity, nucleosome occupancy, consensus binding motifs, and DNA shape, are unable to accurately or fully explain the differential in vivo DNA-binding specificity for 2 paralogous factors expressed at the same time in the same cell. Herein, we dissect the binding determinants for 2 yeast factor paralogues, Cbf1p and Tye7p, and present computational models and in vivo experiments showing that cooperativity and DNA motif information can explain the in vivo specificity for these factors.
Keywords: gene regulation, transcription factor binding, transcription factor cooperativity
Abstract
Eukaryotic cells express transcription factor (TF) paralogues that bind to nearly identical DNA sequences in vitro but bind at different genomic loci and perform different functions in vivo. Predicting how 2 paralogous TFs bind in vivo using DNA sequence alone is an important open problem. Here, we analyzed 2 yeast bHLH TFs, Cbf1p and Tye7p, which have highly similar binding preferences in vitro, yet bind at almost completely nonoverlapping target loci in vivo. We dissected the determinants of specificity for these 2 proteins by making a number of chimeric TFs in which we swapped different domains of Cbf1p and Tye7p and determined the effects on in vivo binding and cellular function. From these experiments, we learned that the Cbf1p dimer achieves its specificity by binding cooperatively with other Cbf1p dimers bound nearby. In contrast, we found that Tye7p achieves its specificity by binding cooperatively with 3 other DNA-binding proteins, Gcr1p, Gcr2p, and Rap1p. Remarkably, most promoters (63%) that are bound by Tye7p do not contain a consensus Tye7p binding site. Using this information, we were able to build simple models to accurately discriminate bound and unbound genomic loci for both Cbf1p and Tye7p. We then successfully reprogrammed the human bHLH NPAS2 to bind Cbf1p in vivo targets and a Tye7p target intergenic region to be bound by Cbf1p. These results demonstrate that the genome-wide binding targets of paralogous TFs can be discriminated using sequence information, and provide lessons about TF specificity that can be applied across the phylogenetic tree.
The first step toward understanding gene regulation is to learn how transcription factors (TFs) bind at specific genomic loci in the cell. Understanding how paralogous TFs achieve their in vivo specificities has proven especially challenging because such factors frequently share highly similar DNA binding-site motifs, yet they are able to specifically discern and bind different target sites in vivo (1–3). This specificity cannot be entirely explained by differences in spatiotemporal expression, since paralogous factors are often present in the cell nucleus at the same time, with presumably the same access to cis-regulatory DNA, but they are still able to bind unique regulatory targets and regulate disparate cellular processes. These challenges make in vivo binding target prediction for paralogous TFs a nontrivial ongoing endeavor.
How do 2 paralogous TFs with nearly identical in vitro DNA binding preferences choose different genomic loci in vivo? Perhaps the most frequently cited explanation draws on the combinatorial nature of eukaryotic TFs and their ability to bind cooperatively to specific cis-regulatory regions. In this model of specificity, direct physical interactions between cofactors allow them to bind at low-affinity sites (4–9). In another example of cooperativity, TFs gain access to their specific binding sites when cofactors mediate DNA bending or repositioning of nearby nucleosomes (10, 11). More recently, it has been shown that the DNA binding preferences of some TFs can be altered upon interaction with cofactor proteins, such that a different “latent” DNA motif is preferred in the presence of these cofactors (2, 12). These examples all represent plausible mechanisms by which paralogous TFs might achieve their specificity, but the goal of explaining the in vivo binding preferences of 2 paralogous TFs from DNA sequence alone has remained elusive.
In this study, we investigated the specificity determinants of the basic−helix–loop–helix (bHLH) TFs Cbf1p and Tye7p in the yeast Saccharomyces cerevisiae. These TFs provide an ideal starting point for the study of paralogous TFs for several reasons. First, they have nearly identical in vitro DNA binding preferences, with a shared consensus CACGTG (Fig. 1A), yet they bind at almost completely nonoverlapping sets of target promoters in vivo (9, 13–15). Second, Cbf1p and Tye7p perform very different cellular functions: Cbf1p is required for chromosome maintenance and growth in the absence of methionine, inducing many of its regulatory targets by recruiting the Met4p activator via a zipper domain located immediately adjacent to its DNA-binding domain (16–19), whereas Tye7p is involved in the regulation of glycolytic genes (20–23). Third, Gordân et al. (13) have carefully investigated the influence of DNA shape on site recognition for these factors. Fourth, unlike many bHLH proteins in higher eukaryotes, these TFs both function as homodimers, negating the need to consider heterodimerization as a contributing factor to binding specificity (14, 19, 22). Lastly, both Cbf1p and Tye7p are constitutively expressed and localized to the nucleus, so both proteins bind their distinct target gene sets during normal, vegetative growth conditions, even though both proteins presumably have the same access to available genomic CACGTG motifs (13, 14, 24).
Fig. 1.
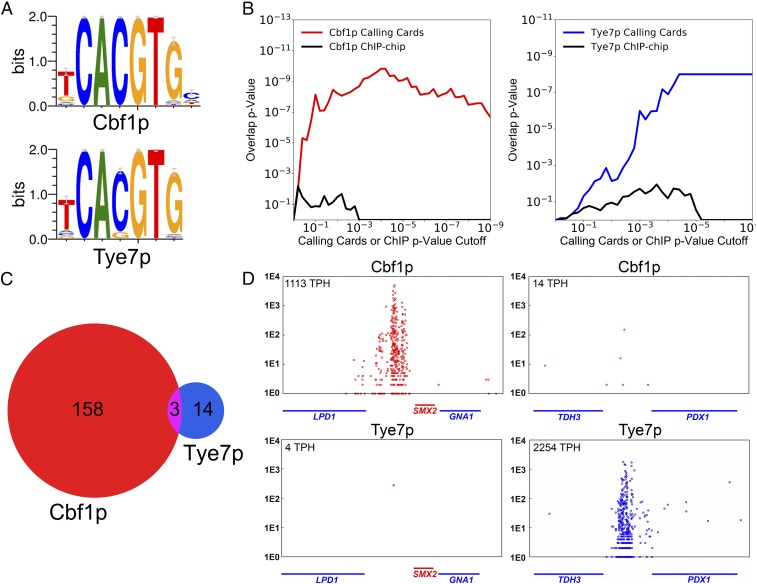
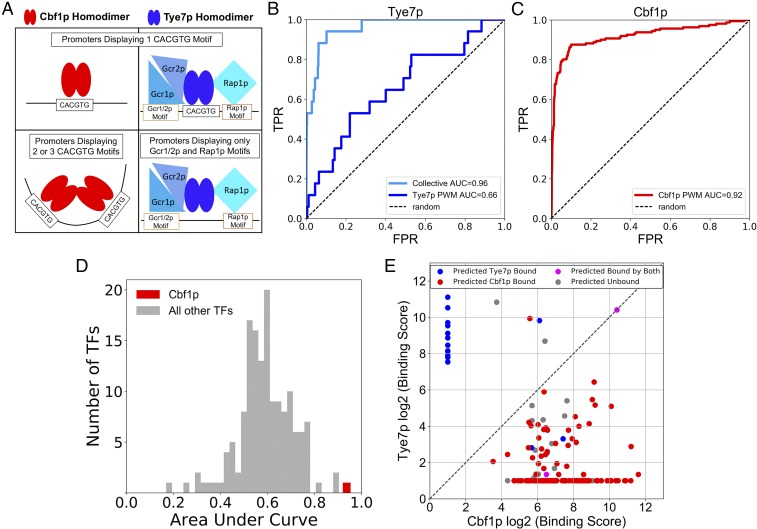
Cbf1p and Tye7p both recognize the CACGTG DNA motif yet bind different genomic targets in vivo. (A) PWMs depicting the consensus binding motif preference of Cbf1p and Tye7p for the canonical E-box sequence CACGTG (47). (B) TF binding targets identified by “calling card” assay have strong overlap with the differentially regulated genes of the corresponding TF deletion strain (31). As the P value cutoff for identification of TF binding targets is lowered (x axis), the P value of the overlap between genome-wide binding targets and differentially regulated genes (y axis) is plotted for Cbf1p (Right) and Tye7p (Left). (C) Cbf1p and Tye7p bind nearly mutually exclusive genomic targets by calling card assay. (D) The binding of Cbf1p and Tye7p at 2 representative loci. The position of red or blue circles on the x axis indicates the genomic location of a factor-directed “calling card” transposon insertion along the chromosome; here, the intergenic regions between LPD1 and SMX2 genes (Left) and TDH3 and PDX1 genes (Right) are shown. The y axis indicates number of sequencing reads supporting a given transposon insertion event. The normalized TPH score of TF binding to the shown locus is given in the top left corner. This “browser shot” presentation is used to illustrate Cbf1p or Tye7p binding at representative genomic loci. (Throughout the figures, the color red is used for Cbf1p, while blue is used for Tye7p when possible.)
To learn how Cbf1p and Tye7p achieve their binding specificities, we made truncated and chimeric TFs and measured their genome-wide binding preferences and cellular functions to uncover which protein domains were important for specificity. This analysis revealed that Cbf1p is the default occupying TF at available CACGTG motifs, in part because of a higher intrinsic affinity for this motif, but also because Cbf1p dimers bind cooperatively with other Cbf1p dimers bound at nearby sites. We show that this homotypic cooperativity depends on Cbf1p′s C-terminal leucine Zipper coil domain (Zip), and that a truncated Cbf1p without this subregion is unable to rescue cbf1Δ. In contrast, we found that Tye7p achieves binding specificity through a genetic interaction with a TF complex consisting of Gcr1p, Gcr2p, and Rap1p. Remarkably, 63% of Tye7p in vivo binding targets do not contain any recognizable CACGTG motif, suggesting that Tye7p binds at most promoters indirectly. Tye7p-bound regulatory regions contain different subsets of Gcr1p, Rap1p, and Tye7p motifs and display no strict rules for motif composition, spacing, or orientation. Thus, Tye7p binds in a manner that is highly reminiscent of the recently described “cooperative collective” model of TF binding (25, 26).
We demonstrated the sufficiency and comprehensiveness of these specificity determinants through 3 computational and experimental avenues. We built computational models that could accurately predict Cbf1p and Tye7p binding in vivo using DNA sequence information alone. We then used the protein features that determine Cbf1p′s specificity to reprogram the human bHLH−PAS TF NPAS2 to complement cbf1Δ. Finally, we converted a Tye7p target promoter to a Cbf1p target promoter by modifying its DNA sequence to nullify Tye7p cofactor binding sites and create Cbf1p binding sites. Together, our results demonstrate that homotypic cooperativity and collective binding are important determinants of bHLH specificity.
Results
Cbf1p and Tye7p Bind Nonoverlapping In Vivo Promoter Targets Despite the Similarity of Their Position-Specific Weight Matrices.
We initiated this study by mapping the in vivo binding of Cbf1p and Tye7p using transposon calling cards, a method designed to measure equilibrium binding (27) and that can detect indirect TF−DNA interactions (28–30). In this method, the TF of interest is C-terminally fused to a short protein tag, which directs insertion of Ty5 retrotransposons near its binding sites. For each assay, a large number of Ty5 insertions are recovered from a population of yeast cells, and their locations are determined via second-generation sequencing, yielding a map of TF binding. As a quantitative measure of binding, we used the number of Ty5 insertion events observed at a promoter in a given experiment, normalized to 100,000 total insertions (transpositions per hundred thousand, TPH). We found that the calling card method is highly reproducible when applied to both Cbf1p and Tye7p (SI Appendix, Fig. S1 A and B), with correlation coefficients between replicates of r = 0.99 and r = 0.99, respectively. Since our strategy to find the specificity determinants for these bHLH proteins involves assaying a number of truncated and chimeric factors expressed from centromeric low-copy plasmids (discussed below), we wanted to ensure that the binding patterns derived from plasmid-borne factors faithfully recapitulate the binding patterns of these factors expressed from endogenous genomic loci. Therefore, we compared binding patterns when Cbf1p and Tye7p were expressed from centromeric plasmids to those obtained when these genes were tagged at their endogenous loci, and we again found a strong concordance (r = 0.93 for Cbf1p, r = 0.88 for Tye7p) (SI Appendix, Fig. S1 C and D). To further verify the accuracy of this system, we compared our calling card data (again when the factors were expressed from centromeric plasmids) to mRNA expression profiles collected from yeast strains in which CBF1 or TYE7 was deleted (31). We observed a high degree of overlap between genes whose promoters were bound by Cbf1p or Tye7p and genes whose expression changed upon deletion of the corresponding factor (Fig. 1B and SI Appendix, Fig. S2). Notably, when Cbf1p or Tye7p binding targets were defined using a previously published chromatin immunoprecipitation (ChIP)-chip dataset (14), concordance with the expression data was reduced (Fig. 1B and SI Appendix, Fig. S2). Together, these results demonstrate that our calling card system reproducibly and accurately measures the in vivo binding of bHLH proteins.
We next analyzed the calling card data for Cbf1p and Tye7p to identify significantly bound intergenic regions (P < 1e-5). We observed little overlap in their sets of binding targets: Only 3 intergenic regions were shared between the 161 Cbf1p and 17 Tye7p targets (Fig. 1C), and one of these is a divergent promoter that regulates both a Cbf1p responsive gene (MET3) and a Tye7p responsive gene (TDH2) (SI Appendix, Fig. S3). Most loci are bound in a nearly binary fashion; for example, the intergenic region between TDH3 and PDX1 is strongly bound by Tye7p, but Cbf1p binding is scarcely above background (Fig. 1 D, Right). In contrast, at the intergenic region between LPD1 and SMX2, Cbf1p binds quite strongly, but no Tye7p binding is observed (Fig. 1 D, Left). This “all-or-none” binding suggests that the specificity of these 2 TFs is not achieved through subtle differences in their DNA binding preferences but instead through substantial differences in affinity at their unique binding targets. The general lack of overlap that we observe between the binding targets of these 2 TFs is consistent with their divergent regulatory functions and with the previous literature (13).
The DNA-Binding Domain of Cbf1p Is Sufficient for Binding and Function, yet No Specific Subregion of the bHLH Is Required for Specificity In Vivo.
We next set out to determine which domains of Cbf1p confer specificity, reasoning that such knowledge could inform interactions with DNA or protein cofactors. A Cbf1p truncation mutant consisting of only the C-terminal bHLH−Zip DNA-binding domain has been found to rescue methionine/cysteine (MET/CYS) prototrophy and chromosome stability in a cbf1Δ strain (19), so we first tested the ability of this truncation mutant (Fig. 2 A, Top) to phenocopy the genome-wide binding pattern of full-length Cbf1p in a cbf1Δ background. Strikingly, Cbf1p bHLH−Zip is able to bind at the targets of full-length Cbf1p (Fig. 2B). From this result, we conclude that the in vivo binding specificity of Cbf1p is encoded solely in its DNA-binding domain.
Fig. 2.
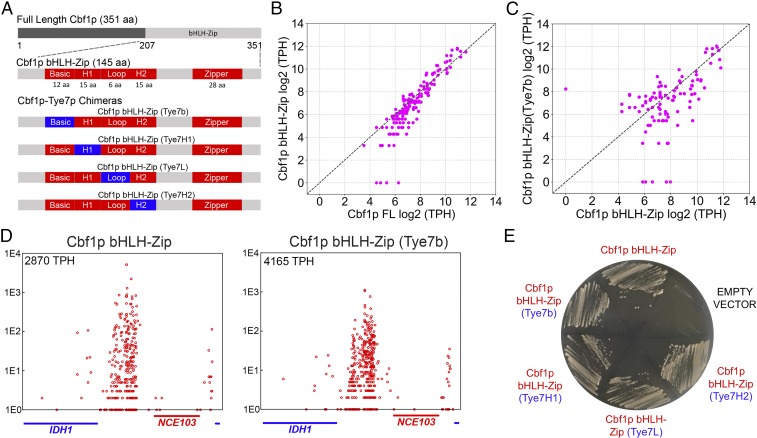
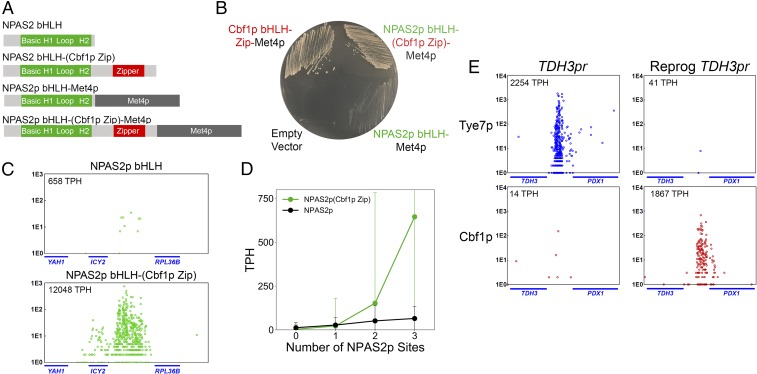
Chimeric Cbf1p bHLH−Zip factors in which individual subregions of the bHLH domain have been replaced with the homologous subregions of Tye7p have wild-type function and genome-wide binding. (A) Protein domain schematics of the Cbf1p, Cbf1p bHLH−Zip, and Cbf1p−Tye7p chimeras used in this study. (B) Cbf1p′s in vivo binding specificity is entirely encoded within its bHLH−Zip domain. The normalized binding (log2 TPH) of full-length Cbf1p is plotted against the binding of Cbf1p bHLH−Zip, as measured by calling card assays. Each point represents a single intergenic region that is significantly bound by either Cbf1p or Cbf1p bHLH−Zip (or both). (C) The Cbf1p bHLH−Zip (Tye7b) chimeric factor binds to nearly all Cbf1p bHLH−Zip targets, with binding measured and displayed as in B. (D) Cbf1p bHLH−Zip (Tye7b) and Cbf1p bHLH−Zip at a representative locus, the intergenic region between the IDH1 and NCE103 genes. (E) Expression of chimeric genes with individual replacement of the basic, helix 1, loop, or helix 2 subregions of Cbf1p bHLH−Zip with those of Tye7p is able to rescue growth of a cbf1Δ strain on agar media lacking MET/CYS.
We then sought to ascertain which subregions of Cbf1p′s DNA-binding domain contribute to its specificity. To do so, we created chimeric proteins in which we switched the basic, helix 1, loop, or helix 2 subregions of Cbf1p bHLH−Zip with the corresponding subregions from Tye7p bHLH (Fig. 2 A, Bottom) and assessed their binding and function. We first analyzed the chimeric factor in which the basic region of Cbf1p bHLH−Zip was switched with the basic region of Tye7p. As with all bHLH proteins, it is the basic subregions of Cbf1p and Tye7p that make specific contacts with DNA bases in the major groove (32, 33). Therefore, if the specificities of these TFs were determined by subtle differences in DNA binding preferences (13, 34), we would expect that this chimeric factor would lose the ability to bind at Cbf1p targets at a minimum, and that it would perhaps spuriously recognize Tye7p targets. However, the in vivo binding of this chimeric protein was very similar to that of Cbf1p, when compared at a genome-wide level (Fig. 2C) or at individual loci, such as the IDH1 promoter (Fig. 2D). Importantly, this chimeric protein was also able to restore MET/CYS prototrophy in a cbf1Δ strain (Fig. 2E), demonstrating that this protein is also functionally indistinguishable from Cbf1p.
Next, we constructed the remaining chimeric factors in which Cbf1p′s helix 1, loop, and helix 2 were exchanged with the corresponding Tye7p subregions (Fig. 2 A, Bottom). We expressed these factors in the cbf1Δ background and assayed for MET/CYS prototrophy and mapped their binding in vivo using transposon calling cards. Remarkably, all of the Cbf1p subregions that we tested could be replaced by their Tye7p counterparts without disrupting MET/CYS prototrophy, whether they are tagged without the calling card Sir4p tag (Fig. 2E) or with it (SI Appendix, Fig. S4). Furthermore, each of these chimeras largely retained Cbf1p′s binding specificity (SI Appendix, Fig. S5), suggesting that this factor’s specificity is not due to unique contacts between the H1, H2, or loop regions and the DNA phosphate backbone. Together, our results suggest that, although the specificity of Cbf1p is entirely encoded in its bHLH−Zip domain, the basic, helix 1, loop, and helix 2 subregions individually contribute little beyond recognition of the CACGTG motif.
The Cbf1p Zip Is Required for Function in an Met4p-Independent Manner and Confers In Vivo Homotypic Cooperative Binding.
Given the apparent dispensability of the basic, helix 1, loop, and helix 2 subregions, we next tested whether the C-terminal Zip domain of Cbf1p is required for specificity. The bHLH−Zip of Cbf1p is known to interact with the Met4p transcriptional activator to regulate MET biosynthetic genes (21). Therefore, to separate the Met4p-recruitment role of the Zip domain from other possible Zip-mediated functions, we engineered a cbf1Δ met4Δ strain and expressed fusions of Cbf1p bHLH−Zip or Cbf1p bHLHΔZip (i.e., no Zip domain) fused in-frame with the Met4p transcriptional activator, as diagrammed in Fig. 3A. On MET/CYS-deficient media, the Cbf1p bHLH−Zip−Met4p fusion protein phenocopies wild-type growth, while Cbf1p bHLH−Met4p displays a strong growth reduction (Fig. 3B). Because the Cbf1p bHLH−Zip and Cbf1p bHLHΔZip proteins are expressed at comparable levels in our system (SI Appendix, Fig. S6), these results indicate that the Zip domain is necessary for Cbf1p to function properly, in a manner independent of its ability to recruit Met4p.
Fig. 3.
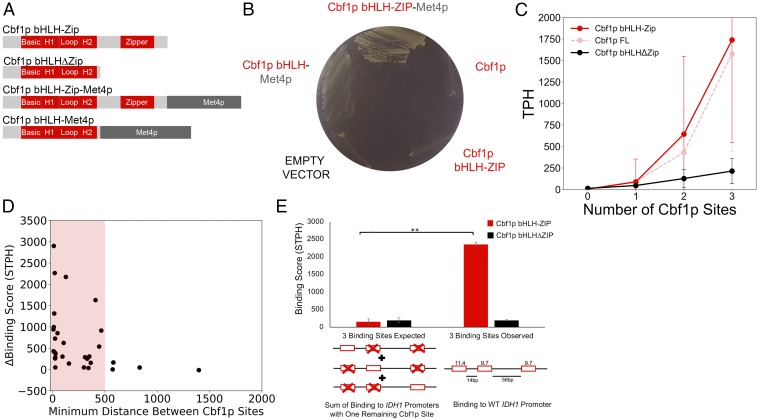
The zipper domain of Cbf1p bHLH−Zip enables homotypic cooperative binding to intergenic regions possessing more than one CACGTG DNA motif and is required for Cbf1p function. (A) Protein domain schematic depicting Cbf1p truncations with and without fusion to Met4p designed to test the function of the Zip domain, Cbf1p′s leucine-repeat zipper coil. (B) In the cbf1Δ met4Δ strain, only a factor possessing both the Met4p transcriptional activator and the Zip of Cbf1p enables wild-type prototrophy on MET/CYS-deficient media, demonstrating that the Zip has a functional role independent of Met4p recruitment. (C) When all yeast intergenic regions are examined for the presence of Cbf1p target motifs meeting a recommended PWM cutoff (47), Cbf1p bHLHΔZip binding increases linearly with the number of CACGTG motifs, while Cbf1p bHLH−Zip binding shows larger increases in a nonlinear fashion. Each point in the graph plots the average normalized binding at all intergenic regions (y axis) with the indicated number of motifs (x axis). Error bars indicate ±1 SD. (D) Cbf1p homotypic cooperative binding is observed when motifs are <500 bp apart. The difference in normalized, background subtracted, binding (STPH) between Cbf1p bHLH−Zip and Cbf1p bHLHΔZip binding to target intergenic regions having ≥2 Cbf1p motifs (y axis) is plotted against the minimum distance between the 2 Cbf1p motifs (x axis). (E) Cbf1p binds with homotypic cooperativity at the IDH1 target promoter (the intergenic region between IDH1 and NCE103 genes) in a Zip-dependent fashion. The sum of truncated factor binding (TPH) to mutated IDH1 promoter intergenic regions displaying only 1 of the 3 endogenous Cbf1p motifs is the “expected” score for binding to the wild-type IDH1 promoter intergenic region, if binding is additive, i.e., not cooperative (Left), vs. the observed binding score of truncated factors to the wild-type 3-site promoter region (Right). Bar height indicates average TPH of 3 independent trials, and error bars indicate ±1 SD. PWM scores for each site are indicated above the wild-type promoter schematic, as well as distance (bp) between sites. **P < 1e-5.
Since Cbf1p′s Zip domain is clearly functionally important, we sought to determine whether it contributes to the binding specificity of this TF. Therefore, we mapped the in vivo binding of Cbf1p bHLH−Zip and Cbf1p bHLHΔZip using transposon calling cards. Although Cbf1p bHLH is bound at many of the same loci as Cbf1p bHLH−Zip (SI Appendix, Fig. S7), there were significant quantitative differences in the binding of these 2 factors. This is shown in Fig. 3C, where we plot the average binding score (TPH) of full-length Cbf1p, Cbf1p bHLH−Zip, and Cbf1p bHLHΔZip at yeast intergenic regions containing 0, 1, 2, or 3 Cbf1p sites (see Materials and Methods). The binding score of Cbf1p bHLHΔZip increases linearly with the number of Cbf1p E-box motifs, whereas the binding scores of either full-length Cbf1p or Cbf1p bHLH−Zip display a sharp nonlinear increase when multiple Cbf1p sites are present (Fig. 3C). This nonlinear increase in binding appears to occur only at intergenic regions with at least 2 Cbf1p sites that are within 500 bp of each other (Fig. 3D). Since Cbf1p bHLH−ZIP and Cbf1p bHLHΔZip proteins are expressed at comparable levels in our cbf1Δ strain (SI Appendix, Fig. S6), the nonlinearity observed for the Zip-containing proteins suggests that this subregion allows Cbf1p homodimers to bind cooperatively with other Cbf1p homodimers bound at nearby sites, in a manner similar to what has been previously observed for the yeast TF Gal4p (35).
To determine whether the Zip subregion is indeed responsible for mediating a cooperative interaction between Cbf1p dimers, we performed a detailed examination of Cbf1p′s binding at the intergenic region between IDH1 and NCE103, a target which harbors 3 Cbf1p E-box motifs. We mutated the Cbf1p binding sites of this intergenic region to all 7 possible combinations of 1, 2, or 3 mutated sites in the cbf1Δ strain and measured the binding of Cbf1p bHLH−Zip and Cbf1p bHLHΔZip to these mutated target loci (see SI Appendix, Fig. S8 A and D for the sequences of the mutated binding targets). If the Cbf1p bHLH−Zip binds to nearby sites independently (i.e., without cooperativity), then we would expect that the binding score observed at the wild-type intergenic region between IDH1 and NCE103 with 3 intact Cbf1p sites would be the sum of the binding scores observed at the mutant regions containing each site in isolation. Instead, we found that Cbf1p bHLH−Zip binding was 15.8-fold higher than would be expected given independent binding (Fig. 3E, P < 0.001). In contrast, binding of the Cbf1p bHLH alone was not statistically different from the value expected under an additive model (197 TPH observed versus 198 TPH expected) (Fig. 3E). Similar results were observed at mutated intergenic regions with 2 Cbf1p binding sites (SI Appendix, Fig. S9). Importantly, cooperative binding of Cbf1p bHLH−ZIP is also observed in a cbf1Δ met4Δ strain (SI Appendix, Fig. S10, Top), demonstrating that Cbf1p′s cooperative binding does not require its known cofactor Met4p. Furthermore, when we expressed Cbf1p bHLHΔZIP from the TEF1 promoter, a strong constitutive yeast promoter (36) (expression quantified by Western blot in SI Appendix, Fig. S6), we still did not observe homotypic cooperative binding of this protein in a cbf1Δ (SI Appendix, Fig. S10, Bottom) or cbf1Δ met4Δ strain (SI Appendix, Fig. S10, Top). Taken together, these findings demonstrate that the Cbf1p Zip subregion mediates a cooperative interaction between Cbf1p dimers bound at nearby sites in the genome. This homotypic interaction results in substantially more Cbf1p binding at yeast promoters with multiple Cbf1p sites, and it is necessary for proper Cbf1p function.
Tye7p Binds via a Cooperative Cofactor Collective.
We next focused on the question of Tye7p′s binding specificity. We initiated our investigation in a manner parallel to that of Cbf1p, by constructing Tye7p truncation and chimeric mutants (diagrammed in SI Appendix, Fig. S11A). As with Cbf1p, we found that Tye7p′s specificity is encoded entirely in its C-terminal DNA-binding domain (SI Appendix, Fig. S11B); therefore, we made chimeric mutants in which we replaced Tye7p′s basic, helix 1, loop, or helix 2 subregions with the homologous subregions of Cbf1p and mapped their binding. We found that the basic region of Tye7p bHLH makes little contribution to the protein’s specificity, as the “basic-swap” chimera binds similarly to full-length Tye7p (SI Appendix, Fig. S11C). However, in contrast with our findings for Cbf1p, we found that swapping the remaining subregions of Tye7p nearly abolishes binding (SI Appendix, Fig. S11D). Since these subregions of the protein do not make base-specific contacts with DNA, these observations led us to hypothesize that Tye7p might function in a complex with other TFs.
To identify which TFs form a complex with Tye7p, we turned to a dataset containing transcriptional profiles for all TF deletion strains in yeast (31). Fourteen of the 17 (82%) Tye7p regulatory targets were down-regulated in the tye7Δ strain, consistent with Tye7p′s known role as a transcriptional activator. Analyzing this dataset further, we found that Gcr1p, Gcr2p, Cst6p, Rap1p, and Sfp1p also activate Tye7p regulatory target genes (P < 1e-4 in each case), suggesting that one or more of these TFs may interact with Tye7p (Fig. 4A). To investigate this possibility, we mapped the in vivo binding of Gcr1p, Gcr2p, and Cst6p using transposon calling cards and analyzed published chromatin endogenous cleavage sequencing (ChEC-seq) and ChIP-seq data for Rap1p (37) and Sfp1p (38). We found that more than 80% of significantly bound Tye7p target regions are strongly bound by the glycolysis regulatory factors Gcr1p and Gcr2p and more than 70% are bound by the general regulatory factor Rap1p (Fig. 4B), while Sfp1p and Cst6p do not significantly cooccupy Tye7p-bound promoters (SI Appendix, Table S1). Furthermore, the binding peaks of Gcr1p, Gcr2p, and Rap1p displayed substantial overlap at individual intergenic regions (SI Appendix, Fig. S12). Gcr2p is known to bind DNA indirectly through its interaction with Gcr1p (39), and the Gcr1/2p complex is also known to physically interact with Rap1p (23, 40–43). Therefore, our results suggested to us that the Gcr1/Gcr2p/Rap1p complex binds cooperatively with Tye7p, an interaction that has not previously been described. To definitively test this hypothesis, we asked whether proper Tye7p binding requires one or more of these cofactors. Since RAP1 is essential for viability (44) and GCR1 deletion results in severe growth defects (45, 46), we assayed Tye7p binding in a gcr2Δ strain and in a wild-type strain. To ensure Tye7p protein levels were equivalent in both strains, we expressed Tye7p from the TEF1 promoter, whose expression is independent of GCR2 in a reporter assay (SI Appendix, Fig. S13). In the absence of the protein product of GCR2, Tye7p binding was severely diminished at nearly all target intergenic regions, such as the intergenic region between TDH3 and PDX1 (Fig. 4 C, Middle). When averaged across all target intergenic regions, Tye7p binding is reduced 8.3-fold (P < 5e-4; Fig. 4D) when Gcr2p is absent, demonstrating that this protein is necessary for Tye7p binding. As an orthogonal test for the dependence of Tye7p binding on Gcr1/2p and Rap1p, we mutated the 2 Rap1p and 2 Gcr1/2p binding sites in the intergenic region between TDH3 and PDX1 (SI Appendix, Fig. S8 B and D). Tye7p binding was completely ablated at this mutated intergenic region (Fig. 4 C, Right, mutated TDH3pr), whereas binding to other target loci was largely unchanged (SI Appendix, Table S2). We conclude that Tye7p binds DNA cooperatively with the Gcr1p/2p Rap1p complex and that this binding is dependent upon the presence of Gcr2p.
Fig. 4.
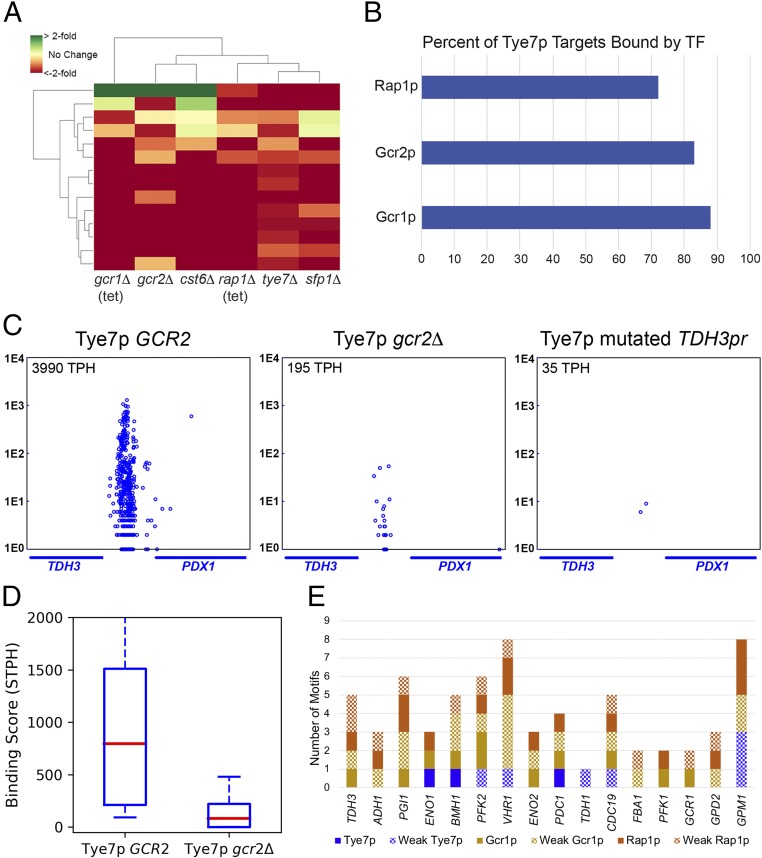
Tye7p achieves in vivo binding specificity through interaction with the Gcr1/2p Rap1p complex. (A) Most genes (82%) whose promoters are bound by Tye7p are down-regulated in a tye7Δ strain, and this set of genes significantly overlaps the down-regulated genes in deletion strains for 5 other TFs. Each column illustrates the transcriptional signature for one deletion strain. [Tet-repressible allele strains were used to reduce expression of the essential genes GCR1 and RAP1 (31).] Each row corresponds to a Tye7p target gene, and the resulting matrix is colored to represent an increase or decrease in gene expression level compared with wild type. (B) Rap1p, Gcr1p, and Gcr2p bind to the majority of Tye7p target loci. Binding targets of Gcr1p, Gcr2p, and Tye7p were determined by calling card assay, while binding targets of Rap1p were previously determined by ChEC-seq (37). (C) Gcr2p is necessary for proper Tye7p binding at the intergenic region between TDH3 and PDX1, as are DNA binding site motifs for Gcr1/2p and Rap1p. (D) Binding of Tye7p is strongly reduced at its targets in the gcr2Δ strain, with an average decrease in binding of 8.3-fold. (E) Motif composition of Tye7p, Gcr1p, and Rap1p binding site motifs for 16 Tye7p targets, using PWMs and scoring cutoffs recommend by ScerTF (47). Weak motifs are defined using a scoring cutoff that is 2 units less than the recommended score. Target gene promoters for Tye7p are indicated along the x axis and were assigned through examination of the transcriptional signature of a tye7Δ strain (31), given significant Tye7p binding to the respective upstream intergenic region. Target gene promoters are presented in order of decreasing Tye7p binding from left to right.
To gain further insight into the nature of the Tye7p Gcr1/2p Rap1p complex, we conducted a binding motif analysis on its intergenic region binding targets. We excluded the divergent promoter regulating MET3 and TDH2, which are Cbf1p- and Tye7p-regulated genes, respectively (SI Appendix, Fig. S3), and identified Tye7p binding motifs using Tye7p′s position-specific weight matrix (PWM). Strikingly, only 3/16 Tye7p targets display a Tye7p motif at the recommended PWM cutoff (47), and 8/16 at a lax cutoff (see Materials and Methods). Thus, half of the intergenic regions where Tye7p is bound do not contain even a weak Tye7p binding motif. However, all but one of its target loci contain binding sites for at least 2 of the 3 DNA-binding proteins in the complex (Fig. 4E). These binding sites tend to occur within 150 bp of each other, but, beyond that, there appear to be no strict rules governing the orientation or spacing of Gcr1p, Tye7p, or Rap1p motifs. The flexible motif composition and grammar that we observe is not consistent with either the classic billboard or enhanceosome models of TF cooperativity, but is instead suggestive of the collective cooperative model recently proposed by Furlong and coworkers (25).
A Model for Cbf1p and Tye7p Binding Specificity.
Our analysis of Cbf1p and Tye7p suggests a simple model (Fig. 5A) to describe how these proteins achieve their in vivo specificities: Cbf1p is the dominant TF at consensus CACGTG sequences by virtue of its intrinsically higher affinity for this sequence (13) and due to a strong homotypic cooperative interaction with other Cbf1p homodimers bound at nearby sites. Tye7p will outcompete Cbf1p at a CACGTG only if there is also a nearby site for one of its cofactors, Gcr1/2p or Rap1p. Furthermore, if there is both a strong Gcr1/2p site and a strong Rap1p site at a promoter, then the E box is dispensable and Tye7p will bind indirectly through these 2 proteins. This framework makes 3 testable predictions. First, by adding information about the binding preferences of Gcr1/2p and Rap1p, we should be able to discriminate Tye7p-bound intergenic regions from unbound intergenic regions significantly better than if we only consider Tye7p′s PWM. Second, since Cbf1p dominates at CACGTGs, its PWM alone should have strong predictive power. This would be atypical, as a PWM does not accurately predict the in vivo binding pattern for the majority of eukaryotic TFs (48–50). Third, by applying a simple decision tree based on the rules described above, we should be able to discriminate promoters bound by Cbf1p from those bound by Tye7p.
Fig. 5.
The in vivo binding patterns of Cbf1p and Tye7p can be predicted by computational models only examining proximal DNA sequence information. (A) Model of the determinants of in vivo binding specificity for Cbf1p and Tye7p (see text for detail). (B) Tye7p-bound promoter regions can be distinguished from unbound regions by combining PWM score information for Gcr1p, Rap1p, and Tye7p motifs. ROC displaying the ability of Tye7p PWM score to differentiate between bound and unbound promoters (true promoter targets identified by “calling card” assay) (dark blue line) is shown compared with a model incorporating collective TF PWM scores for each promoter in the genome (light blue line). (C) The PWM score of the highest-scoring Cbf1p motif in a given genomic promoter region differentiates Cbf1p-bound and unbound promoters. (D) Of all yeast TFs having PWMs, Cbf1p PWM motif score is the best-performing PWM in the prediction of in vivo binding targets. (E) A decision tree based on specificity determinants accurately distinguishes in vivo genomic Cbf1p and Tye7p binding targets. Each point represents a Cbf1p- or Tye7p-bound intergenic region, and the point color indicates predicted TF binding at that locus, based on PWM motif scores for Cbf1p and Tye7p collective members (see text for details).
To test the first prediction, we used Tye7p′s PWM to discriminate a set of Tye7p-bound intergenic regions from the set of unbound promoters (as determined by calling cards) and plotted the results using a receiver operator curve (ROC) (51) (Fig. 5B). Tye7p′s PWM alone performed significantly better than chance, but its ability to discriminate bound and unbound promoters was modest (area under the ROC [AUROC] = 0.66). However, when we added information about Gcr1/2p′s and Rap1p′s binding preferences by taking the sum of the highest-scoring PWMs for each of these 3 factors within a 150-bp window (see Materials and Methods), the performance improved dramatically (AUROC = 0.96). Together, our results confirm that, by utilizing information about the binding preferences of Tye7p′s cofactors, we explain the in vivo binding preferences of this factor.
We next gauged the ability of Cbf1p′s PWM to identify the in vivo targets of this factor. Strikingly, a ROC analysis with Cbf1p′s PWM alone achieved an AUROC of 0.93, suggesting that the presence of a single high-scoring Cbf1p binding motif is highly predictive of Cbf1p′s in vivo binding (Fig. 5C). To put this performance in context, we performed a similar analysis for an additional 155 S. cerevisiae TFs, using an optimized set of PWMs (47) to predict in vivo binding as measured previously in a comprehensive in vivo ChIP-chip dataset (14). None of the TFs we analyzed achieved an AUROC higher than was observed for Cbf1p, confirming our original hypothesis that Cbf1p′s PWM would be a uniquely powerful predictor (Fig. 5D). We also confirmed that Cbf1p′s in vivo binding levels could be more accurately predicted using a more sophisticated model that included homotypic cooperative interactions, at the cost of including additional parameters (SI Appendix, Table S3).
Finally, we created a decision tree to distinguish promoters bound by Cbf1p from those bound by Tye7p. This decision tree is based on the simple model described above (Fig. 5A). Intergenic regions with a single E box (a site that can be bound by either Tye7p or Cbf1p) but no nearby Rap1p or Gcr1p sites are classified as Cbf1p targets. Intergenic regions with motifs for 2 of 3 TFs in the Tye7p complex within 150 bp are classified as Tye7p targets, and intergenic regions with 2 or more E boxes are classified as Cbf1p targets (see Materials and Methods). Intergenic regions that do not satisfy any of these rules are classified as unbound, and intergenic regions that have 2 distinct sequence regions that can be separately assigned to Tye7p and Cbf1p are classified as bound by both TFs. This decision tree was used to classify the set of intergenic regions bound by Cbf1p or Tye7p, and the results are shown in Fig. 5E. Each data point represents a yeast intergenic region, and the x coordinate gives the Cbf1p in vivo binding score on a log2 scale, while the y coordinate gives the Tye7p in vivo binding score on a log2 scale. Thus, data points along the x axis represent intergenic regions bound exclusively by Cbf1p in vivo, while those along the y axis represent intergenic regions bound exclusively by Tye7p. The points are then colored according to their predicted classification: red for Cbf1p-bound, blue for Tye7p-bound, gray for unbound, and pink for bound by both factors. Fig. 5E demonstrates that a simple decision tree can accurately distinguish Cbf1p- and Tye7p-bound intergenic regions, especially those that are exclusively bound by one factor or the other (i.e., the points along the x and y axes).
Functional Reprogramming of a Human bHLH TF.
Our model predicts that the specificity of Cbf1p is determined by its intrinsic affinity for the CACGTG E box and by a homotypic cooperative interaction governed by its C-terminal Zip domain. If these 2 factors are indeed sufficient for Cbf1p′s in vivo specificity, then the fusion of Cbf1p′s Zip to the C terminus of a human bHLH that binds at CACGTG sequences in vitro should endow the factor with Cbf1p′s binding pattern and should also functionally complement the yeast protein if fused to an activation domain. The human bHLH NPAS2 (neuronal PAS domain-containing protein 2) is a member of the bHLH−PAS TF family and binds CACGTG motifs but is not known to utilize homotypic cooperativity (52, 53). We added the Met4p transcriptional activator to the C terminus of either the NPAS2 bHLH alone or the NPAS2 bHLH−(Cbf1p Zip) fusion (diagrammed in Fig. 6A) and tested for rescue of the cbf1Δ met4Δ strain. As shown in Fig. 6B, the NPAS2 bHLH−(Cbf1p Zip) protein functionally rescues MET/CYS prototrophy, whereas the NPAS2 bHLH alone does not. Binding analysis reveals that NPAS2 bHLH−(Cbf1p Zip) but not NPAS2 bHLH binds to individual Cbf1p targets such as the ICY2 promoter (Fig. 6C). Furthermore, a genome-wide analysis of NPAS2 bHLH−(Cbf1p Zip) at intergenic regions containing 1, 2, or 3 motifs represented by the NPAS2 PWM reveals that, as predicted, the Cbf1p Zip confers homotypic cooperative binding to NPAS2 bHLH (Fig. 6D). Importantly, NPAS2 bHLH−(Cbf1p Zip) and NPAS2 bHLH alone display equivalent expression in the cbf1Δ met4Δ strain (SI Appendix, Fig. S14). From this result, we conclude that CACGTG site recognition and homotypic cooperativity are sufficient for Cbf1p′s in vivo binding specificity and function.
Fig. 6.
The human bHLH−PAS NPAS2 can be reprogrammed to phenocopy Cbf1p bHLH−Zip in vivo binding specificity and function. (A) Protein domain schematic of truncated NPAS2 factors, with and without fusion to Met4p, designed to achieve Cbf1p-like genome-wide binding and function. (See Materials and Methods for details.) (B) NPAS2p bHLH−(Cbf1p Zip) fused to Met4p functionally complements the cbf1Δ met4Δ strain on MET/CYS-deficient media, whereas NPAS2p bHLH fused to Met4p does not. (C) NPAS2p bHLH−(Cbf1p Zip) binds to the Cbf1p target intergenic region between ICY2 and RPL36B (Bottom), while NPAS2p lacking the Cbf1p Zip displays substantially reduced binding (Top), as measured by calling card assays. (D) Binding of NPAS2p bHLH−(Cbf1p Zip) increases in a nonlinear fashion when 2 or 3 NPAS2 binding site motifs are within a single intergenic region. (E) Cbf1p, but not Tye7p, binds to a “reprogrammed” Tye7p target intergenic region (between TDH3 and PDX1) in which Rap1p and Gcr1/2p sites were mutated and 3 CACGTG motifs were added.
Binding Specificity Reprogramming of Cbf1p and Tye7p Target Intergenic Regions via DNA Sequence Determinants.
Lastly, we sought to test the predictive power of our model by reprograming a Tye7p target intergenic region so that it would instead be bound by Cbf1p. To do so, we mutated 2 Rap1p sites and 2 Gcr1p sites to prevent binding by the Tye7p Gcr1/2p Rap1p complex and modified the surrounding sequences to include 3 CACGTG motifs. To “reprogram” this intergenic region, we used only base pair substitutions, as opposed to indels (see SI Appendix, Fig. S8 C and D for “reprogrammed” TDH3 promoter sequence). We observe strong binding of Cbf1p, but not Tye7p, to our “reprogrammed” TDH3 promoter (the intergenic region between TDH3 and PDX1), which is the converse of the natural binding pattern (Fig. 6E). (Tye7p binding to other target intergenic regions was largely unchanged in the “reprogrammed” TDH3 promoter strain, as shown in SI Appendix, Table S2.) This reprogramming further exemplifies a detailed understanding of Cbf1p and Tye7p binding specificity determinants.
Discussion
How do TFs choose their target sites in vivo? While PWMs provide invaluable information, knowledge of motif alone fails to predict in vivo binding for most TFs (48, 49, 54). This problem is exacerbated for paralogous TFs, which add an additional layer of complexity to in vivo binding site prediction because they typically share remarkably similar PWMs but bind at distinct target sets. In this study, we dissected the specificity determinants of the paralogous yeast bHLH proteins Cbf1p and Tye7p, both of which bind at CACGTG-containing sequences in vitro. We found that Cbf1p achieves its specificity through a high intrinsic affinity for the E box and by binding cooperatively with other Cbf1p homodimers bound at nearby sites. In contrast, we found that Tye7p achieves its specificity by binding in a “cooperative collective” with 3 other factors, Gcr1p, Gcr2p, and Rap1p. Using this knowledge, we formulated a simple conceptual model to describe how these TFs bind in vivo and demonstrated its predictive power through 3 distinct computational and experimental avenues. Together, our results show that homotypic cooperativity and collective binding are key determinants of bHLH specificity.
What is the mechanism by which Cbf1p′s Zip domain confers homotypic cooperativity? Previous studies examining the structure of USF1, a Cbf1p ortholog, suggest that the Cbf1p Zip may tetramerize via a coiled-coil interaction to mediate the observed homotypic cooperativity (32, 55). Because we did not directly measure this putative coiled-coil interaction, it is also possible that the homotypic cooperativity we observe is mediated by another protein that bridges 2 Cbf1p dimers. Cbf1p′s Zip domain is not only required for proper binding, but it is also clearly necessary for proper function of the TF, since we found that MET/CYS prototrophy requires an intact Zip domain. Furthermore, this functional requirement is independent of the transcriptional activator, MET4p, whose recruitment to MET promoters is an additional Zip function. We therefore conclude that the homotypic interaction mediated by the Zipper domain is essential for Cbf1p′s function. Relatively few examples of a functional phenotypic requirement for homotypic cooperative TF binding have been found, with the most salient example being the necessity of homotypic cooperative binding by Bicoid in the embryonic patterning of Drosophila (56).
Our main findings regarding the determinants of Cbf1p′s specificity are compatible with earlier functional studies. For example, previous work has found that Cbf1p can act as a pioneer factor (57), consistent with our observation that Cbf1p′s motif predicts in vivo binding more accurately than other TF motifs. Siggers et al. (12) previously found that the RYAAT motif is required for the transcriptional activation of Cbf1-dependent sulfur metabolism genes. Together with our results, this suggests a model where Cbf1p binds E boxes independently of the RYAAT motif at most target promoters, and then efficiently recruits Met4p and Met28p to the subset of bound loci that encode the RYAAT motif. Such a model is also consistent with the previous observation that Cbf1p can act as either an activator or a repressor (58, 59).
Although Tye7p has never been previously shown to bind cooperatively with the Gcr1p/Gcr2p/Rap1p complex, previous studies on the functions of these proteins are completely consistent with such an interaction, which could be the result of direct protein−protein contacts between Tye7p and this complex or could be mediated by an adaptor protein. Tye7p displays genetic interactions with the Gcr1p and Gcr2p (23, 42); Gcr1p and Gcr2p were already known to function in a TF complex consisting of Rap1p and the nuclear pore protein Nup84p (41, 43). Furthermore, the binding of Gcr1p and Rap1p to promoters of glycolytic enzyme genes has been shown to be combinatorial in nature, with the binding of Gcr1p dependent upon the presence of bound Rap1p (40). Also, classical studies of elements in the glycolysis enzyme gene promoters revealed transcriptional synergy of Gcr1/2p and Rap1p sites in the TDH3 and PGK1 gene promoters (60–62). Thus, our finding that Gcr1p, Gcr2p, Rap1p, and Tye7p bind at highly overlapping sets of binding targets agrees well with the existing literature on these proteins.
It is notable that the majority of Tye7p′s in vivo targets (63%) lack a recognizable CACGTG consensus sequence, suggesting that Tye7p binds at most promoters indirectly. Consistent with this hypothesis, nearly all Tye7p-bound intergenic regions contain moderately strong motifs for at least 2 of the 3 DNA-binding proteins in the Tye7p/Gcr1p/Gcr2p/Rap1p complex, but these motifs display no strict rules for motif composition, spacing, or orientation. Taken together, these observations suggest that Tye7p does not conform to either of the 2 classical models of TF cooperativity—the enhanceosome model or the “billboard” model. In the enhanceosome model, TFs bind in a highly cooperative fashion to elements possessing the motifs for all of the factors in well-ordered spacing and orientation (63). In the “billboard” model, all of the TF motifs for each factor are again present in the regulatory region, but not all motifs must be bound to achieve transcriptional activity, since factors binding to the “billboard” can influence gene expression either independently or cooperatively (64). Notably, in both of these models, a TF binding motif is required at each regulatory element where the TF acts (26). However, in the case of Tye7p, we observe many yeast intergenic regions that are bound but do not encode the corresponding motif (Fig. 4E). Thus, Tye7p′s binding is highly reminiscent of the recently proposed cooperative collective model for TF binding in higher eukaryotes (25, 26). In this model, members of a TF complex bind cooperatively to enhancer elements without a fixed motif composition or strict grammar, with optimal synergistic output depending only on the presence of most TFs of the complex. Motifs for all of the occupying TFs need not be present on any given cis-regulatory module, suggesting variability and flexibility in protein−protein and protein−DNA interactions (25, 65–67). The Tye7p/Gcr1p/Gcr2p/Rap1p regulatory complex represents an excellent model system to ascertain the rules and principles that govern this poorly understood model of cooperative binding.
The work presented herein demonstrates that homotypic cooperativity and collective binding are utilized by paralogous TFs to achieve their binding specificities, complementing and extending previous investigations into the roles of DNA shape and differences in intrinsic binding affinities (1, 13, 68, 69). Since most TFs expressed in higher eukaryotes such as humans and mice are members of large paralogous gene families (70), we expect that these mechanisms will be broadly employed throughout the phylogenetic tree.
Materials and Methods
Referenced details of the materials and methods, including yeast strains and culturing conditions, plasmid construction and design, yeast calling card assay, paired-end sequence map back for Ty5 insertion site identification, identification of target intergenic (promoter) regions, Western blotting, B-galactosidase reporter assays, PWMs, ROCs, binding target prediction decision tree, and further bioinformatic analysis are provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank the personnel at the DNA Sequencing Innovation Lab and High Throughput Computing Facility at The Edison Family Center for Genome Sciences and Systems Biology of Washington University in St. Louis for their sequencing and computational expertise, respectively. We thank Nikhil Mitra for writing the Shea−Ackers model. We are also grateful to Michael Brent, Barak Cohen, Gary Stormo, and Heather True for helpful discussion and reagents. This work was supported by National Institute of Health Grants R01GM123203, R01NS076993, U01MH109133, and RF1MH117070.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data discussed in this publication have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE134427). Custom scripts and code are available at GitLab, https://gitlab.com/rob.mitra/shively_2019.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1818015116/-/DCSupplemental.
References
- 1.Shen N., et al. , Divergence in DNA specificity among paralogous transcription factors contributes to their differential in vivo binding. Cell Syst. 6, 470–483.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Slattery M., et al. , Cofactor binding evokes latent differences in DNA binding specificity between Hox proteins. Cell 147, 1270–1282 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Voth W. P., et al. , Forkhead proteins control the outcome of transcription factor binding by antiactivation. EMBO J. 26, 4324–4334 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Val S., et al. , Combinatorial regulation of endothelial gene expression by ets and forkhead transcription factors. Cell 135, 1053–1064 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferraris L., et al. , Combinatorial binding of transcription factors in the pluripotency control regions of the genome. Genome Res. 21, 1055–1064 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fong A. P., et al. , Conversion of MyoD to a neurogenic factor: Binding site specificity determines lineage. Cell Rep. 10, 1937–1946 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frey F., et al. , Molecular basis of PRC1 targeting to Polycomb response elements by PhoRC. Genes Dev. 30, 1116–1127 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hollenhorst P. C., et al. , DNA specificity determinants associate with distinct transcription factor functions. PLoS Genet. 5, e1000778 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou X., O’Shea E. K., Integrated approaches reveal determinants of genome-wide binding and function of the transcription factor Pho4. Mol. Cell 42, 826–836 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buck M. J., Lieb J. D., A chromatin-mediated mechanism for specification of conditional transcription factor targets. Nat. Genet. 38, 1446–1451 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Falvo J. V., Thanos D., Maniatis T., Reversal of intrinsic DNA bends in the IFN beta gene enhancer by transcription factors and the architectural protein HMG I(Y). Cell 83, 1101–1111 (1995). [DOI] [PubMed] [Google Scholar]
- 12.Siggers T., Duyzend M. H., Reddy J., Khan S., Bulyk M. L., Non-DNA-binding cofactors enhance DNA-binding specificity of a transcriptional regulatory complex. Mol. Syst. Biol. 7, 555 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gordân R., et al. , Genomic regions flanking E-box binding sites influence DNA binding specificity of bHLH transcription factors through DNA shape. Cell Rep. 3, 1093–1104 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harbison C. T., et al. , Transcriptional regulatory code of a eukaryotic genome. Nature 431, 99–104 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rossi M. J., Lai W. K. M., Pugh B. F., Genome-wide determinants of sequence-specific DNA binding of general regulatory factors. Genome Res. 28, 497–508 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baker R. E., Masison D. C., Isolation of the gene encoding the Saccharomyces cerevisiae centromere-binding protein CP1. Mol. Cell. Biol. 10, 2458–2467 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cai M., Davis R. W., Yeast centromere binding protein CBF1, of the helix-loop-helix protein family, is required for chromosome stability and methionine prototrophy. Cell 61, 437–446 (1990). [DOI] [PubMed] [Google Scholar]
- 18.Cai M. J., Davis R. W., Purification of a yeast centromere-binding protein that is able to distinguish single base-pair mutations in its recognition site. Mol. Cell. Biol. 9, 2544–2550 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mellor J., et al. , CPF1, a yeast protein which functions in centromeres and promoters. EMBO J. 9, 4017–4026 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benanti J. A., Cheung S. K., Brady M. C., Toczyski D. P., A proteomic screen reveals SCFGrr1 targets that regulate the glycolytic-gluconeogenic switch. Nat. Cell Biol. 9, 1184–1191 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Kuras L., Cherest H., Surdin-Kerjan Y., Thomas D., A heteromeric complex containing the centromere binding factor 1 and two basic leucine zipper factors, Met4 and Met28, mediates the transcription activation of yeast sulfur metabolism. EMBO J. 15, 2519–2529 (1996). [PMC free article] [PubMed] [Google Scholar]
- 22.Löhning C., Ciriacy M., The TYE7 gene of Saccharomyces cerevisiae encodes a putative bHLH-LZ transcription factor required for Ty1-mediated gene expression. Yeast 10, 1329–1339 (1994). [DOI] [PubMed] [Google Scholar]
- 23.Nishi K., et al. , The GCR1 requirement for yeast glycolytic gene expression is suppressed by dominant mutations in the SGC1 gene, which encodes a novel basic-helix-loop-helix protein. Mol. Cell. Biol. 15, 2646–2653 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huh W.-K., et al. , Global analysis of protein localization in budding yeast. Nature 425, 686–691 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Junion G., et al. , A transcription factor collective defines cardiac cell fate and reflects lineage history. Cell 148, 473–486 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Spitz F., Furlong E. E. M., Transcription factors: From enhancer binding to developmental control. Nat. Rev. Genet. 13, 613–626 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Wang Y., Guo L., Golding I., Cox E. C., Ong N. P., Quantitative transcription factor binding kinetics at the single-molecule level. Biophys. J. 96, 609–620 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang H., Heinz M. E., Crosby S. D., Johnston M., Mitra R. D., ‘Calling Cards’ method for high-throughput identification of targets of yeast DNA-binding proteins. Nat. Protoc. 3, 1569–1577 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Wang H., Johnston M., Mitra R. D., Calling cards for DNA-binding proteins. Genome Res. 17, 1202–1209 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang H., Mayhew D., Chen X., Johnston M., Mitra R. D., Calling Cards enable multiplexed identification of the genomic targets of DNA-binding proteins. Genome Res. 21, 748–755 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu Z., Killion P. J., Iyer V. R., Genetic reconstruction of a functional transcriptional regulatory network. Nat. Genet. 39, 683–687 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Ferré-D’Amaré A. R., Pognonec P., Roeder R. G., Burley S. K., Structure and function of the b/HLH/Z domain of USF. EMBO J. 13, 180–189 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones S., An overview of the basic helix-loop-helix proteins. Genome Biol. 5, 226 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maerkl S. J., Quake S. R., A systems approach to measuring the binding energy landscapes of transcription factors. Science 315, 233–237 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Giniger E., Ptashne M., Cooperative DNA binding of the yeast transcriptional activator GAL4. Proc. Natl. Acad. Sci. U.S.A. 85, 382–386 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Partow S., Siewers V., Bjørn S., Nielsen J., Maury J., Characterization of different promoters for designing a new expression vector in Saccharomyces cerevisiae. Yeast 27, 955–964 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Zentner G. E., Kasinathan S., Xin B., Rohs R., Henikoff S., ChEC-seq kinetics discriminates transcription factor binding sites by DNA sequence and shape in vivo. Nat. Commun. 6, 8733 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Albert B., et al. , Sfp1 regulates transcriptional networks driving cell growth and division through multiple promoter-binding modes. Genes Dev. 33, 288–293 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deminoff S. J., Santangelo G. M., Rap1p requires Gcr1p and Gcr2p homodimers to activate ribosomal protein and glycolytic genes, respectively. Genetics 158, 133–143 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Drazinic C. M., Smerage J. B., López M. C., Baker H. V., Activation mechanism of the multifunctional transcription factor repressor-activator protein 1 (Rap1p). Mol. Cell. Biol. 16, 3187–3196 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Menon B. B., et al. , Reverse recruitment: The Nup84 nuclear pore subcomplex mediates Rap1/Gcr1/Gcr2 transcriptional activation. Proc. Natl. Acad. Sci. U.S.A. 102, 5749–5754 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sato T., et al. , The E-box DNA binding protein Sgc1p suppresses the gcr2 mutation, which is involved in transcriptional activation of glycolytic genes in Saccharomyces cerevisiae. FEBS Lett. 463, 307–311 (1999). [DOI] [PubMed] [Google Scholar]
- 43.Tornow J., Zeng X., Gao W., Santangelo G. M., GCR1, a transcriptional activator in Saccharomyces cerevisiae, complexes with RAP1 and can function without its DNA binding domain. EMBO J. 12, 2431–2437 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Conrad M. N., Wright J. H., Wolf A. J., Zakian V. A., RAP1 protein interacts with yeast telomeres in vivo: Overproduction alters telomere structure and decreases chromosome stability. Cell 63, 739–750 (1990). [DOI] [PubMed] [Google Scholar]
- 45.Baker H. V., Glycolytic gene expression in Saccharomyces cerevisiae: Nucleotide sequence of GCR1, null mutants, and evidence for expression. Mol. Cell. Biol. 6, 3774–3784 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clifton D., Fraenkel D. G., The gcr (glycolysis regulation) mutation of Saccharomyces cerevisiae. J. Biol. Chem. 256, 13074–13078 (1981). [PubMed] [Google Scholar]
- 47.Spivak A. T., Stormo G. D., ScerTF: A comprehensive database of benchmarked position weight matrices for Saccharomyces species. Nucleic Acids Res. 40, D162–D168 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arvey A., Agius P., Noble W. S., Leslie C., Sequence and chromatin determinants of cell-type-specific transcription factor binding. Genome Res. 22, 1723–1734 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kumar S., Bucher P., Predicting transcription factor site occupancy using DNA sequence intrinsic and cell-type specific chromatin features. BMC Bioinformatics 17 (suppl. 1), 4 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Orenstein Y., Shamir R., A comparative analysis of transcription factor binding models learned from PBM, HT-SELEX and ChIP data. Nucleic Acids Res. 42, e63 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fawcett T., An introduction to ROC analysis. Pattern Recognit. Lett. 27, 861–874 (2006). [Google Scholar]
- 52.Khan A., et al. , JASPAR 2018: Update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 46, D260–D266 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou Y. D., et al. , Molecular characterization of two mammalian bHLH-PAS domain proteins selectively expressed in the central nervous system. Proc. Natl. Acad. Sci. U.S.A. 94, 713–718 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu X., Lee C.-K., Granek J. A., Clarke N. D., Lieb J. D., Whole-genome comparison of Leu3 binding in vitro and in vivo reveals the importance of nucleosome occupancy in target site selection. Genome Res. 16, 1517–1528 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lamber E. P., Wilmanns M., Svergun D. I., Low resolution structural models of the basic helix-loop-helix leucine zipper domain of upstream stimulatory factor 1 and its complexes with DNA from small angle X-ray scattering data. Biophys. J. 94, 193–197 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lebrecht D., et al. , Bicoid cooperative DNA binding is critical for embryonic patterning in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 102, 13176–13181 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yan C., Chen H., Bai L., Systematic study of nucleosome-displacing factors in budding yeast. Mol. Cell 71, 294–305.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee T. A., et al. , Dissection of combinatorial control by the Met4 transcriptional complex. Mol. Biol. Cell 21, 456–469 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McIsaac R. S., Petti A. A., Bussemaker H. J., Botstein D., Perturbation-based analysis and modeling of combinatorial regulation in the yeast sulfur assimilation pathway. Mol. Biol. Cell 23, 2993–3007 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bitter G. A., Chang K. K., Egan K. M., A multi-component upstream activation sequence of the Saccharomyces cerevisiae glyceraldehyde-3-phosphate dehydrogenase gene promoter. Mol. Gen. Genet. 231, 22–32 (1991). [DOI] [PubMed] [Google Scholar]
- 61.Chambers A., Packham E. A., Graham I. R., Control of glycolytic gene expression in the budding yeast (Saccharomyces cerevisiae). Curr. Genet. 29, 1–9 (1995). [DOI] [PubMed] [Google Scholar]
- 62.Stanway C. A., Chambers A., Kingsman A. J., Kingsman S. M., Characterization of the transcriptional potency of sub-elements of the UAS of the yeast PGK gene in a PGK mini-promoter. Nucleic Acids Res. 17, 9205–9218 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thanos D., Maniatis T., Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell 83, 1091–1100 (1995). [DOI] [PubMed] [Google Scholar]
- 64.Arnosti D. N., Kulkarni M. M., Transcriptional enhancers: Intelligent enhanceosomes or flexible billboards? J. Cell. Biochem. 94, 890–898 (2005). [DOI] [PubMed] [Google Scholar]
- 65.Doitsidou M., et al. , A combinatorial regulatory signature controls terminal differentiation of the dopaminergic nervous system in C. elegans. Genes Dev. 27, 1391–1405 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Uhl J. D., Cook T. A., Gebelein B., Comparing anterior and posterior Hox complex formation reveals guidelines for predicting cis-regulatory elements. Dev. Biol. 343, 154–166 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Uhl J. D., Zandvakili A., Gebelein B., A hox transcription factor collective binds a highly conserved distal-less cis-regulatory module to generate robust transcriptional outcomes. PLoS Genet. 12, e1005981 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Abe N., et al. , Deconvolving the recognition of DNA shape from sequence. Cell 161, 307–318 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rohs R., et al. , The role of DNA shape in protein-DNA recognition. Nature 461, 1248–1253 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wingender E., Schoeps T., Dönitz J., TFClass: An expandable hierarchical classification of human transcription factors. Nucleic Acids Res. 41, D165–D170 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.