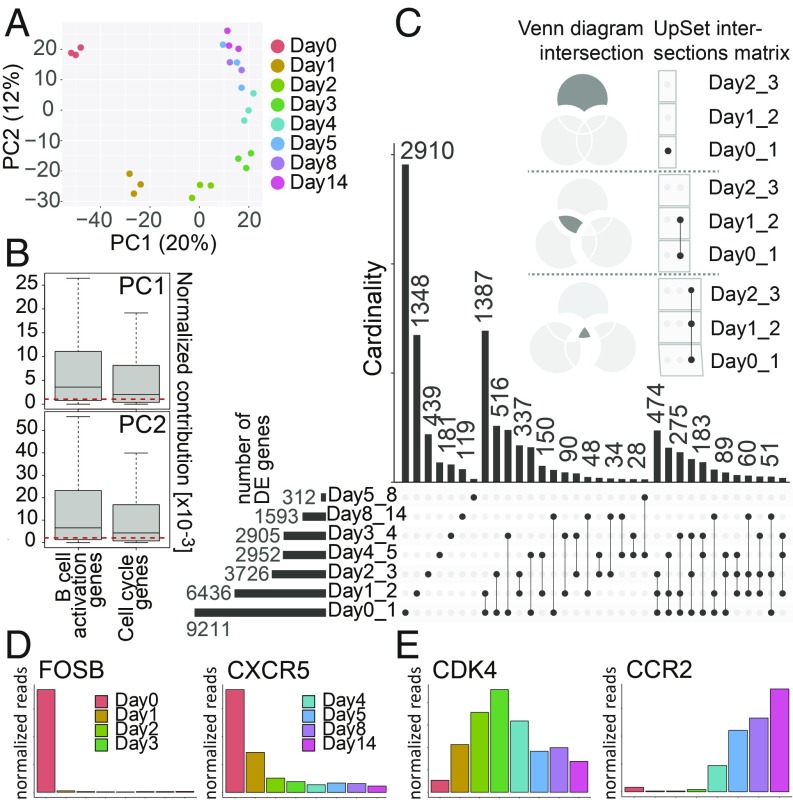
Fig. 2.
Analysis of the time-resolved RNA-seq data. (A) PCA of the samples from sequencing libraries at the given time points postinfection. The daily samples are represented by 3 biological replicates with the exception of day 8 (n = 2). The samples are shown as a function of PC1 and PC2. The x and y axes show the percentages of variance explained by PC1 and PC2. (B) The normalized contributions of cell cycle annotated genes and genes marking any of the stages of B cell activation (Fig. 5A) to PC1 and PC2, as plotted in A, are shown. The genes that are annotated as both cell cycle and B cell activation genes were removed from the analysis. The red dashed lines indicate the median normalized contributions calculated on all genes. The contributions of both cell cycle genes and B cell activation genes are statistically significantly higher than expected (Wilcoxon rank-sum test; P values for cell cycle genes are 0.0002 and 8 × 10−11 for PC1 and PC2, respectively; P values for B cell activation genes are <2.2 × 10−16 for both PC1 and PC2). The y axes provide the scaled values (×10−3) of their normalized contributions. (C) The UpSet plot visualizes intersecting sets of DE genes at different time points after EBV infection. In the upper part of this panel, 3 Venn diagrams serve as examples to illustrate the syntax of the UpSet diagram. The single dot and dots with interconnecting vertical lines form the intersection matrix. Paired intersections are depicted as black dots; gray dots indicate the sets that are not part of the intersection. Black lines connecting 2 or more black dots indicate which sets form the intersections. In the lower part, groups of DE genes, obtained from the pairwise comparison (performed by DESeq2) (SI Appendix, Materials and Methods) as indicated (Day0_1 = day 0 vs. day 1, etc.) form 7 sets in the UpSet matrix, which are depicted as black horizontal bars with “Number of DE genes” to indicate the set sizes (ranging from 312 to 9,211). Each row represents a matrix set. The columns show the intersections depicted as areas in the schematic Venn diagram as explained in upper part of this panel. The heights of the black columns indicate the cardinality (number of elements) in the different intersections. (D) The bar plots show the time-resolved expression of 2 selected genes, which displayed high expression in noninfected cells or only in cells very early after EBV infection. These genes are among those with the highest loadings on PC1 and/or PC2. The x axis displays days postinfection, whereas the y axis depicts normalized read counts averaged over different biological replicates. (E) Mean expression of 2 genes that contributed to cell cycle progression and B cell activation as function of time after infection. The indicated genes were barely expressed in noninfected cells but became activated at different time points after EBV infection.