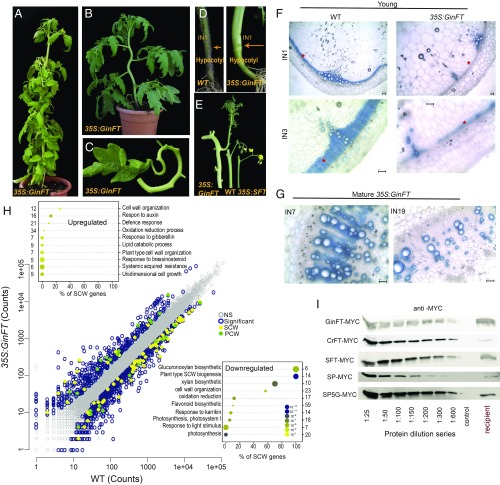
Fig. 4.
The gymnosperm GinFT gene antagonizes florigen, suppresses SCWB, and prolongs vascular maturation. (A–E) Characterization of the GinFT developmental syndrome in tomato. (A) A mature 35S:GinFT-MYC transgenic plant. The inward curling of stems and leaves is age-dependent. (B) A convoluted stem from a 4-wk-old 35S:GinFT plant. (C) The petiole and rachis of an adult 35S:GinFT leaf. Lateral leaflets have been removed for demonstration. (D) Hypocotyls of 35S:GinFT plants undergo differential radial constriction. Arrows mark the hypocotyl stem junctions. (E) Size comparisons of inflorescence-bearing stems. (Left) 35S:GinFT; (Center) WT; (Right) 35S:SFT. (F and G) High expression levels of GinFT delays lignification of IF fibers in young (F) and mature (G) stems. (F) 30-d-old WT and 35S:GinFT plants. (Upper) Internode 1. (Lower) Internode 3 at higher magnification. (G) 75-d-old 35S:GinFT plants, internodes 7 and 19. Note the accumulation of nonlignified xylem fibers. TBO staining for lignin. (H) SCWB genes are significantly down-regulated in 35S:GinFT stems. The scatterplot shows expression levels of 35S:GinFT vs. WT. Colored circles represent 1,122 DEGs (Padj < 0.1; |FC ≥2|). (Insets) Top 10 GO terms in up-regulated (Top) and down-regulated (Bottom) DEGs. (I) CETS proteins of gymnosperms conserve mobility potential. Protein blots show a dilution series of MYC-tagged proteins from the indicated CETS donor leaves, along with the undiluted mobile CETS proteins from the leaves of the corresponding recipient shoots. *IF zones. (Scale bars: 100 mm.)