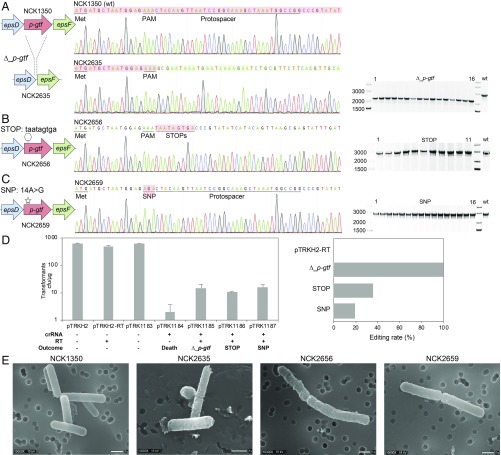
Fig. 4.
Diversity of genome editing outcomes achieved by repurposing the endogenous type I-E system in L. crispatus NCK1350. Different editing strategies can be achieved based on the repair template cloned in the targeting plasmid (SI Appendix, Figs. S1 and S2A). (A) Deletion of 643 bp in the exopolysaccharide p-gtf gene, with the chromatogram showing the sequence of the NCK1350 wild-type strain (wt) and the deletion mutant NCK2635. (B) Insertion of stop codons while deleting the protospacer region in the p-gtf gene to generate the mutant NCK2656 (eps15_16::taatagtga). (C) Single base editing performed as single base substitution to alter the PAM sequence (14A > G), creating a missense mutation (K5R) in the derivative mutant NCK2659. (D) Transformation efficiencies and editing rates (%). cfu, colony-forming unit. (E) Scanning electron microscopy of the wild-type strain L. crispatus NCK1350 and the derivative mutants NCK2635, NCK2656, and NCK2659 harboring a deletion, interruption, or single base substitution in the exopolysaccharide p-gtf gene, respectively. (Magnification: 10,000× to 13,000×.) (Scale bar: 1 μm.) Images courtesy of North Carolina State University/Claudio Hidalgo-Cantabrana and Valerie Lapham.