Figure 8.
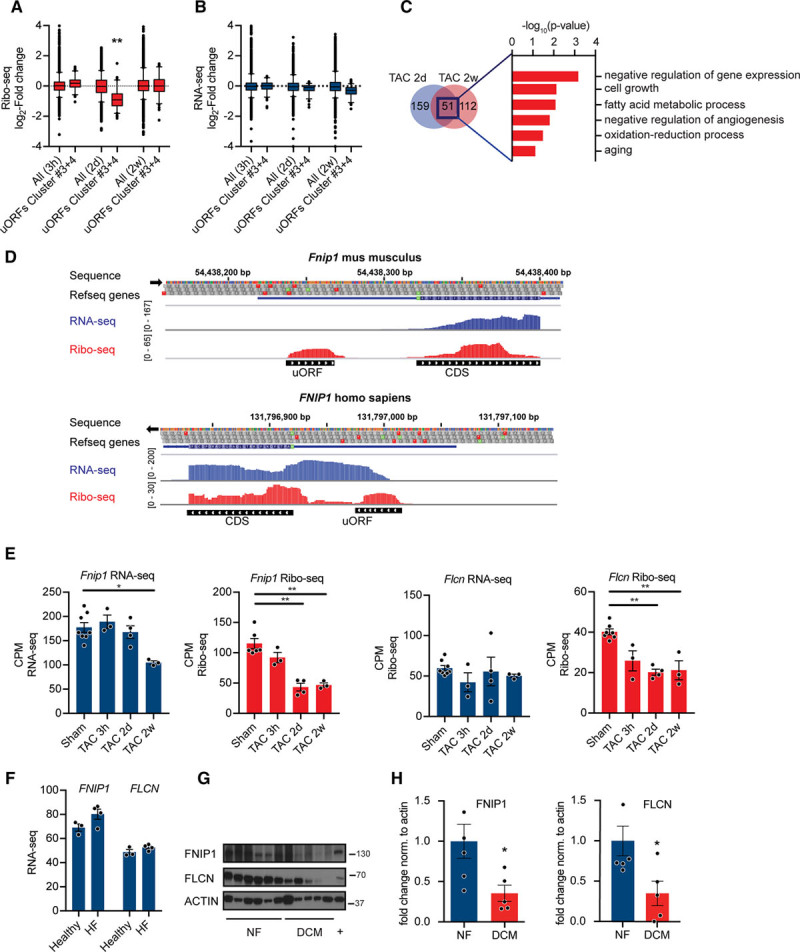
Regulation of gene expression by upstream open reading frame (uORFs). A, Ribo-seq counts per million (CPM) for transcripts in cluster 4 at different time points after transverse aortic constriction (TAC) surgery. N≥3 for each time point; **P<0.01 Kruskal-Wallis test. Whiskers represent 5% to 95% CI. B, RNA-seq CPM for transcripts in cluster 4 at different time points after TAC surgery. N≥3 for each time point. C, Venn diagram showing overlap between significantly less translated transcripts containing a regulatory uORF at 2d and 2w after TAC. Enrichment of gene ontology terms (biological process) in differentially expressed transcripts after TAC surgery with uORFs. −log10 P values, Fisher exact test. D, Ribo-seq (red), RNA-seq (blue), and coverage plots for the H. sapiens and M. musculus genome loci containing FNIP1 (folliculin-interacting protein 1) with reads mapped to the 5′ UTR. E, Ribo-seq CPM and RNA-seq CPM for Fnip1 and Flcn (folliculin) at different time points after TAC surgery. *False discovery rate (FDR) <0.05, **FDR <0.01. F, RNA-seq counts for FNIP1 and FLCN in human heart samples. N=4. G, Immunoblots and (H) quantification for FNIP1 and FLCN in human heart samples confirming decreased expression in dilated cardiomyopathies (DCM). P<0.05; n≥5. Two-tailed Student unpaired t test. CDS indicates coding sequence.
