Table 1.
Simulation results for our generalized meta-analysis estimators in the logistic regression setting; estimated standard deviations were obtained by taking averages over simulated datasets and were used to construct 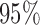 confidence intervals, whose coverage rates and average lengths are reported
confidence intervals, whose coverage rates and average lengths are reported
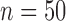
|
|
Bias | SD (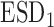 , , 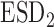 ) ) |
RMSE | CR | AL |
|---|---|---|---|---|---|---|
| genmeta.0 |
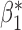
|
0.010 | 0.161 (0.161, 0.162) | 0.161 | 0.968, 0.964 | 0.642, 0.636 |
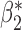
|
0.005 | 0.110 (0.111, 0.108) | 0.110 | 0.958, 0.960 | 0.434, 0.423 | |
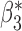
|
 0.001 0.001 |
0.138 (0.143, 0.142) | 0.138 | 0.963, 0.964 | 0.559, 0.556 | |
| genmeta.1 |
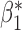
|
0.005 | 0.117 (0.116, 0.110) | 0.117 | 0.976, 0.966 | 0.455, 0.433 |
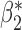
|
 0.003 0.003 |
0.101 (0.105, 0.099) | 0.101 | 0.964, 0.955 | 0.411, 0.386 | |
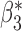
|
0.001 | 0.099 (0.102, 0.097) | 0.099 | 0.973, 0.961 | 0.402, 0.381 | |
| genmeta.2 |
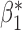
|
0.007 | 0.115 (0.116, 0.111) | 0.115 | 0.971, 0.964 | 0.455, 0.435 |
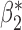
|
 0.003 0.003 |
0.102 (0.105, 0.099) | 0.102 | 0.960, 0.959 | 0.413, 0.388 | |
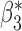
|
0.003 | 0.098 (0.103, 0.098) | 0.098 | 0.957, 0.957 | 0.403, 0.383 |
SD, standard deviation; ESD , estimated standard deviation using the reference sample; ESD
, estimated standard deviation using the reference sample; ESD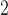 , estimated standard deviation using the covariance estimates of reduced model parameters from the studies; RMSE, square root of mean square error; CR, coverage rate of 95% confidence intervals; AL, average length of 95% confidence intervals.
, estimated standard deviation using the covariance estimates of reduced model parameters from the studies; RMSE, square root of mean square error; CR, coverage rate of 95% confidence intervals; AL, average length of 95% confidence intervals.
