Abstract
Nuclear ribosomal DNA (nrDNA) sequences are widely used in the molecular classification of fungi. Previous phylogenetic studies of highly-valued traditional Chinese medicinal fungus Ophiocordyceps sinensis were mostly based on 18S and internal transcribed spacer (ITS) regions (ITS1, 5.8S and ITS2) of nrDNA. However, the disparity manifest in the low sequences identities between different O. sinensis isolates has led to argumentative hypotheses for this phenomenon, such as the “species complex” or “cryptic species” hypotheses. In the present study, four types of nrDNA (GC, AT-1, AT-2, and T) were identified using four primer pairs to amplify the nrDNA of six O. sinensis isolates. We demonstrate that each O. sinensis isolate contained two types of nrDNA, the omnipresent GC-type and a coexistent type alternating between the remaining three. This crucial discovery challenges the established notion of one type of nrDNA per species. We therefore propose that the composition of nrDNA types should be taken into consideration in studies of fungal genetics and classification.
Keywords: Ophiocordyceps sinensis, nrDNA, species complex, cryptic species
1. Introduction
Ophiocordyceps sinensis (Berk.) Sacc. is a typical entomogenous fungus found in southwestern China. It grows parasitically on the larvae of Lepidoptera, particularly those belonging to the genus Hepialus. In Asia, O. sinensis is considered to be a valuable traditional fungus that has several medicinal effects [1].
Using phylogenetic trees based on the ITS region of nrDNA sequences (ITS1, 5.8S and ITS2 nrDNA sequences), Kinjo and Zang [2] suggested that the 17 collected O. sinensis isolates from 11 southwestern China localities could be divided into two subgroups. Stensrud et al. [3] analyzed ITS region nrDNA variation among 71 sequences of O. sinensis available in the EMBL/GenBank databases, and suggested that O. sinensis isolates could be divided into three subgroups and that “cryptic species” existed within the O. sinensis grouping. Whether it is the “subgroup” concept or “cryptic species” concept, low sequences identities are not normally observed in the Cordyceps/Ophiocordyceps genus.
In the present study, we designed four primer pairs to amplify the partial 18S and ITS region nrDNA sequences from six O. sinensis isolates. We have demonstrated that a single O. sinensis isolate could simultaneously have two types of nrDNA sequences.
2. Materials and Methods
2.1. Fungal Specimens and Strains
The specimens and strains used in this study are listed in Table 1. Strains were cultured in 250 mL potato dextrose broth (PDB; DIFCO, Detroit, MI, USA) in 500 mL flasks agitated at 100 rpm at 10 °C. The mycelia were harvested after 8 weeks and washed with sterile water. All specimens and mycelia were then lyophilized and stored at −20 °C prior to further analysis.
Table 1. Ophiocordyceps sinensis isolates examined in this study.
| Isolate No. | Tissue | Locality | Primer pair | Product size (bp) | Accession No. | nrDNA Type |
| Cs7528A-0901 | Stroma | Tibet, China | NS5/GCITS4 | 1219 | AM931048 | GC |
| NS5/AT12ITS4 | 1223 | AM931055 | AT-2 | |||
| Korea-0901 | Stroma | Qinghai, China | NS5/GCITS4 | 1219 | AM931050 | GC |
| TITS5b/TITS4 | 477 | AM931059 | T | |||
| W1023-0901 | Stroma | Sichuan, China | NS5/GCITS4 | 1219 | AM931049 | GC |
| TITS5b/TITS4 | 477 | AM931060 | T | |||
| RS2-0901 | Mycelia | Tibet, China | NS5/GCITS4 | 1219 | AM931051 | GC |
| NS5/AT12ITS4 | 1219 | AM931057 | AT-1 | |||
| RS3-0901 | Mycelia | Tibet, China | NS5/GCITS4 | 1219 | AM931052 | GC |
| NS5/AT12ITS4 | 1223 | AM931056 | AT-2 | |||
| RS4-2-0901 | Mycelia | Tibet, China | NS5/GCITS4 | 1219 | AM931053 | GC |
| TITS5b/TITS4 | 477 | AM931058 | T |
2.2. DNA Preparation
DNA was isolated as described by Moncalvo et al. [4]. Briefly, the ground sample (60 mg) was transferred to a 1.5 mL microcentrifuge tube containing 600 µL of lysis buffer (50 mM Tris-HCl, 50 mM EDTA, 3% SDS, and 1% 2-mercaptoethanol at pH 7.2). The tube was incubated in a water bath at 65 °C for 1 h, and the aqueous phase was then extracted twice using 600 µL of PCI (phenol/chloroform/isoamyl alcohol = 25:24:1; Sigma Co., Saint Louis, MO, USA). After extraction, the aqueous phase was transferred to a new tube, and the precipitated DNA was mixed with 0.1 volumes 3 M sodium acetate to 0.6 volumes isopropanol. The DNA was centrifuged at 15 000× g for 5 min, washed twice with cold 70% ethanol and dried for 30 min in a vacuum oven at 37 °C. The remaining DNA pellet was then resuspended in 100 µL of TE buffer (10 mM Tris-HCl and 1 mM EDTA; pH 8.0) containing 2 µL of RNase (500 µg/mL; Roche Applied Science Co., Mannheim, Germany) and incubated in a water bath at 37 °C for 1 h. After the addition of 100 µL of chloroform, the aqueous phase was transferred directly into a new tube. DNA was precipitated using 0.1 volumes 3 M sodium acetate to 0.6 volumes isopropanol and then again centrifuged at 15 000× g for 5 min. The DNA was resuspended in 100 µL sterile water and stored at −20 °C.
2.3. PCR Amplification
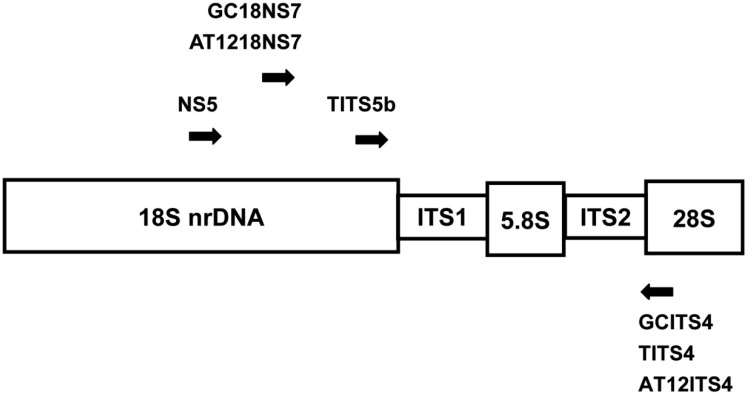
The primers were designed according to White et al. [5] and Kinjo and Zang [2]. The primers sequences and locations used in the present study are given in Table 2 and Figure 1. Amplifications were carried out using a Perkin-Elmer thermocycler 480 (Applied Biosystems, Foster City, CA, USA) in 25 µL reaction mixture containing 50 ng template DNA, 2.5 µL 10× PCR buffer (ProTech Professional Technical Services, Inc., Pittsburgh, PA, USA), 0.2 mM of each dNTP, 0.5 µM of each primer, and 0.625 U of super taq DNA polymerase (ProTech Professional Technical Services, Inc., Pittsburgh, PA, USA). Amplifications were performed using the following PCR program: initial denaturation at 96 °C for 2 min, followed by 35 cycles at 96 °C for 45 s, 54 °C for 45 s, 72 °C for 2 min, and a final extension step at 72 °C for 10 min. A negative control (dsH2O) was included.
Table 2. Primers used in this study [2],[5].
| Primer | Primer sequence (5′→3′) |
| NS5 | AACTAAAGGAATTGACGGAAG |
| GCITS4 | ATCCGAGGTCAACTGGAGGGTGTG |
| AT12ITS4 | GCTTCCGGTGCGAGGTTCTCGGTG |
| TITS4 | AGTGCGAGGTTCTTAGTAAGCTATTGCG |
| TITS5b | TCGAGTTACCACTCCTAAACCCCCTGC |
| GC18NS7 | TCCGGCAGTGCGCCGGCTTC |
| AT1218NS7 | CGTACTACTCTAGTAGTACGCCGGCTTG |
Figure 1. Locations on nrDNA of PCR primers designed in the present study. Arrowheads represent the 3′ end of each primer.
Gel electrophoresis of PCR products was performed on a 1.2% agarose gel containing 2 mL ethidium bromide per 100 mL TAE buffer (0.02 M Tris base, 0.02 M acetate and 0.0005 M EDTA, AMRESCO Co., Solon, Ohio, USA). Gels were visualized using an UV-trans-illuminator.
2.4. DNA Sequencing and Phylogenetic Analyses
PCR-amplified products were sequenced by the Mission Biotech Company (Taipei, Taiwan). The sequences were analyzed in an auto sequencer (Applied Biosystems, Foster City, CA, USA) using a terminator cycler sequencing ready reaction kit (Applied Biosystems, USA). Additional sequences that did not originate from the sequencing of these 12 isolates were downloaded from NCBI. Completed sequences were imported into the program BioEdit sequence alignment editor version 7.0.9.0. [6], and aligned using the CLUSTALW [7] option. Phylogenetic trees were constructed using maximum likelihood analysis in the phylogenetic analysis using parsimony 4.0 program (PAUP 4.0, http://paup.csit.fsu.edu/).
2.5. Different Types of nrDNA Copy Number Ratio Analyses
The different types of nrDNA were amplified 10 to 35 cycles, respectively, to estimate copy number ratio. The forward primers GC18NS7 and AT1218NS7 and reverse primers GCITS4, AT12ITS4, and TITS4 (Table 2, Figure 1) pairs were used to amplify ITS1, 5.8S, and ITS2 nrDNA.
3. Results
Primer pair NS5/GCITS4 amplified sequences from all six isolates, which yielded products of 1219 bp. Primer pair NS5/AT12ITS4 amplified sequences from isolates Cs7528A-0901 and RS3-0901, yielding products of 1223 bp. NS5/TITS4 was unable to amplify the sequence of any isolate. We replaced this pair with TITS5b/TITS4, and by doing so we were able to successfully amplify ITS region sequences from isolates Korea-0901, W1023-0901, and RS4-2-0901. However, the resulting products were smaller at 477 bp. The GenBank accession numbers of the amplified products are shown in Table 1.
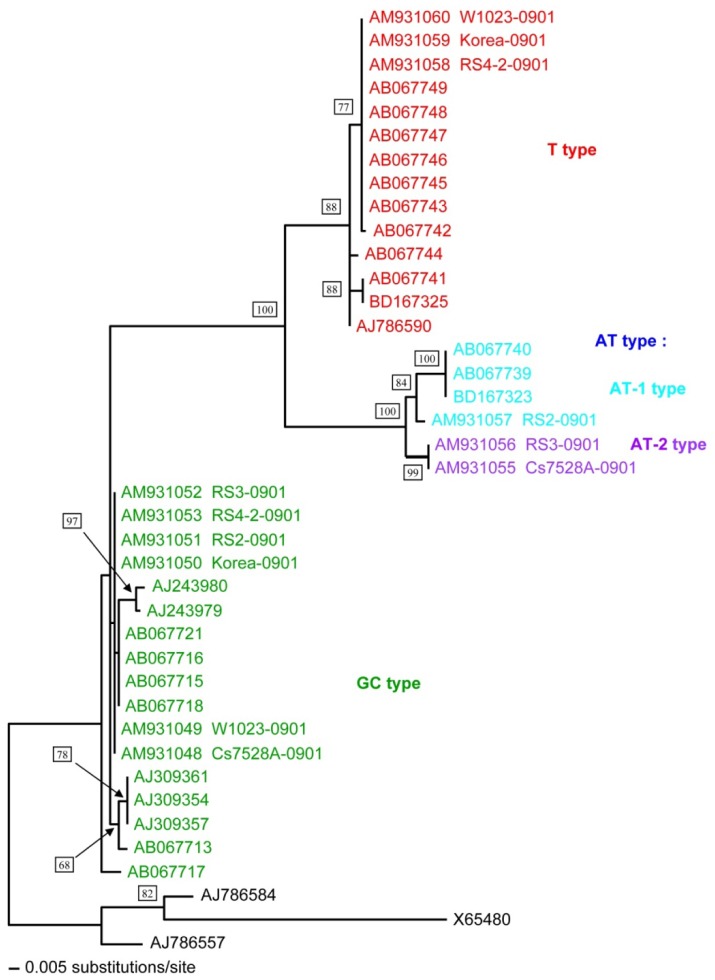
Based on the ITS region sequences phylogenetic tree (Figure 2), O. sinensis could be divided into three types: GC-type (GC content 64.6%), T-type (GC content 54.4%) and AT-type (GC content 51.4%; this could be further divided into AT-1 and AT-2 types). The sequence identities between each type were between 85.1% and 96.7% (Table 3).
Figure 2. Phylogenetic tree resulting from the Maximum Likelihood Method (ML) of the nrDNA ITS1, 5.8S, ITS2 region sequences of Ophiocordyceps sinensis and other related fungi. Bootstrap percentage values of ML are shown at each branch.
Table 3. Sequence identities between different types of ITS region nrDNA in Ophiocordyceps sinensis.
| nrDNA types | T | AT-1 | AT-2 | GC |
| T | 1.000 | 0.898 | 0.891 | 0.894 |
| AT-1 | 1.000 | 0.967 | 0.862 | |
| AT-2 | 1.000 | 0.851 | ||
| GC | 1.000 |
Crucially, we found that each O. sinensis isolate simultaneously had two types of ITS region nrDNA sequences (Table 1, Figure 3): the GC-type sequence and one other sequence type. Analogous results showed that a single O. sinensis isolate had two types of 18S nrDNA sequence (partial) simultaneously. Isolate Cs7528A-0901 contained both the GC-type and AT-type sequences, RS2-0901 contained the GC-type and AT-type sequences, and RS3-0901 contained the GC-type and AT-type sequences. The sequence identities between each type were between 84.7% and 98.6% (Table 4).
Figure 3. The nrDNA 18S and ITS region types of Ophiocordyceps sinensis isolates. The green band indicates the GC-type sequence, the purple band indicates the AT-2 type, and the blue band indicates the AT-1 type. The red band, in which was discovered only the ITS region, indicates the T type.
Table 4. Sequence identities between different types of 18S nrDNA (partial) in Ophiocordyceps sinensis.
| nrDNA types | AT-1 | AT-2 | GC |
| AT-1 | 1.000 | 0.986 | 0.849 |
| AT-2 | 1.000 | 0.847 | |
| GC | 1.000 |
The different types of nrDNA were amplified 10 to 35 cycles to estimate copy number ratio. Only primer pairs GC18NS7/GCITS4 (GC-type) and AT1218NS7/AT12ITS4 (AT-type) could successfully amplify ITS1, 5.8S, and ITS2 nrDNA. Other primer pairs, e.g., GC18NS7/AT12ITS4, GC18NS7/TITS4, AT1218NS7/GCITS4, and AT1218NS7/TITS4 failed. For example, in strain RS2-0901, the AT-type first appeared after the 20th cycle and the GC-type first appeared after the 16th cycle, indicating that the GC-type was 16-fold greater than the AT-type.
4. Discussion
nrDNA sequences are widely used in the molecular classification of fungi. The primers used in such analyses are mainly designed based on the nrDNA sequences of Saccharomyces cerevisiae [5]. By virtue of primer specificity, usually unique-type sequences are amplified and used for phylogenetic analysis. Otherwise, the absence of reference sequences restricts the design of primers, and other types of sequences are difficult to amplify. Therefore, a single species has traditionally been considered to have one nrDNA sequence. According to this concept, most earlier molecular researchers amplified only the GC-type nrDNA sequences and consequently inferred a low sequence divergence within O. sinensis [8]–[15]. When different types of nrDNA were amplified in earlier molecular studies, they were interpreted as being subgroups of a “species complex” [2] or as “cryptic species” [3].
In the present study, the O. sinensis isolates were indistinguishable from each other on the basis of morphological features. However, each had the constituent GC-type nrDNA as well as other types of nrDNA. This result demonstrated that classification of O. sinensis (and other fungal species) based on nrDNA sequences should ideally take into consideration the composition of nrDNA types.
An outstanding question, however, is why isolates Korea-0901, W1023-0901, and RS4-2-0901, which have the T-type ITS region nrDNA sequence, did not have corresponding T-type 18S nrDNA sequences. There are two possible explanations for this disparity. The T-type ITS region nrDNA sequence may correspond to the GC-type 18S nrDNA sequence. Alternatively, the T-type ITS region nrDNA sequence may correspond to the T-type 18S nrDNA sequence, which we have been unable to amplify. Since no amplification products were obtained using the primer pair NS5 (GC-type 18S nrDNA forward primer)/TITS4 (T-type ITS region nrDNA reverse primer), the second explanation may be more plausible.
There are also literary records of single species having more than one nrDNA sequences, including the Ascomycetes Fusarium graminearum, Gibberella fujikuroi [16],[17], and Neurospora [18], the Basidiomycetes Trichaptum abietinum [19], various plant species [20], the tiger beetle Cicindela dorsalis [21], and the nematode species Meloidogyne hapla and M. chitwoodi [22]. Notably, the variation of nrDNA in the listed citations only occurs in locations that do not affect mature rRNA function. For example, the two types of nrDNA in F. graminearum and G. fujikuroi [16],[17] occur mainly in the ITS2 region, and in T. abietinum [19], tri-type nrDNA occurred mainly in the ITS1 region, after the formation of pre-rRNA would have been removed. Thus, rRNA and ribosome composition are not affected.
In the present study, we found not only the variation of nrDNA in ITS1 and ITS2, but also that there were multiple types of rRNA and ribosomes in single O. sinensis in the functional 18S and 5.8S nrDNA regions. The differences in ribosomes may lead to differences in their synthetic proteins, and may also lead to regionally-sourced O. sinensis having different medicinal effects. In addition, intra-individual variation in nrDNA is common, and different copies with different GC-content are frequent. Such AT-rich copies are usually considered non-functional pseudogenes [23]. The low GC content characteristic of many non-recombining genomes may be the result of three processes: (1) a prevailing AT mutational bias; (2) random fixation of the most common types of mutation; and (3) the absence of the GC-biased gene conversion, which, in recombining organisms, permits the reversal of the most common types of mutation [24]. According to this theory, the AT-type and T-type of O. sinensis may have experienced a shift in life history towards a prevailingly non-recombining clonal life style, such as the anamorph stage of life cycle [3].
Another hypothesis raised to account for the AT bias observed in obligate biotrophs is that competition for metabolic resources may exist between host and parasite [25]. In O. sinensis, this could be due to situations such as a shift towards an even more obligate biotrophic life style in AT-type and T-type, which is the anamorph stage [3]. Further research, including a larger pool of molecular and ecological data on O. sinensis isolates in nature and from culture, will be required to resolve the above hypotheses.
Acknowledgments
This study was supported by a grant (Grant No. DOH-91-TD-1163) from the Department of Health, Executive Yuan, Taiwan. We are grateful to Emeritus Professor Yong Hang of Cornell University, Ithaca, New York, USA for reviewing and giving critical comments regarding this manuscript.
Abbreviations
- ITS
internal transcribed spacer
- nrDNA
nuclear ribosomal DNA
- PCR
polymerase chain reaction
Footnotes
Conflict of Interest: The authors have no conflict of interest to declare.
References
- 1.Chen CS, Huang CT, Hseu RS. Identification of Cordyceps sinensis by 18S nrDNA Sequencing and Characterization of Fermented Products in Taiwan. Food Biotechnol. 2009;23:1–9. [Google Scholar]
- 2.Kinjo N, Zang M. Morphological and phylogenetic studies on Cordyceps sinensis distributed in southwestern China. Mycoscience. 2001;42:567–574. [Google Scholar]
- 3.Stensrud Ø, Schumacher T, Shalchian-Tabrizi K, et al. Accelerated nrDNA evolution and profound AT bias in the medicinal fungus Cordyceps sinensis. Mycol Res. 2007;111:409–415. doi: 10.1016/j.mycres.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 4.Moncalvo JM, Wang HH, Hseu RS. Gene phylogeny of the Ganoderma lucidum complex based on ribosomal DNA sequences. Comparison with traditional taxonomic characters. Mycol Res. 1995;99:1489–1499. [Google Scholar]
- 5.White TJ, Bruns T, Lee S, et al. Amplification and direct sequencing of fungal ribosomal DNA for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, et al., editors. PCR protocols: a guide to methods and applications. San Diego, California: Academic Press, Inc; 1990. pp. 315–322. [Google Scholar]
- 6.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 7.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen CS, Hseu RS. Differentiation of Cordyceps sinensis (Berk.) Sacc. specimen using restriction fragment length polymorphism of 18S rRNA gene. J Chin Agri Chem Soc. 1999;37:533–545. [Google Scholar]
- 9.Chen CS, Hseu RS. Differentiation of Cordyceps sinensis (Berk.) Sacc. with 18S rRNA Gene Sequences. Taiwan J Agri Chem Food Sci. 2002;40:219–225. [Google Scholar]
- 10.Chen CS, Ling CH, Hseu RS. Differentiation of Cordyceps sinensis (Berk.) Sacc. with Internal Transcribed Spacer1, 5.8S and Internal Transcribed Spacer 2 rRNA Gene Sequences. Taiwan J Agri Chem Food Sci. 2004;42:147–153. [Google Scholar]
- 11.Chen YQ, Hu B, Xu F, et al. Genetic variation of Cordyceps sinensis, a fruit-body-producing entomopathogenic species from different geographical regions in China. FEMS Microbiol Lett. 2004;230:153–158. doi: 10.1016/S0378-1097(03)00889-9. [DOI] [PubMed] [Google Scholar]
- 12.Chen YQ, Wang N, Qu L, et al. Determination of the anamorph of Cordyceps sinensis inferred from the analysis of the ribosomal DNA internal transcribed spacers and 5.8S rDNA. Biochem Syst Ecol. 2001;29:597–607. doi: 10.1016/s0305-1978(00)00100-9. [DOI] [PubMed] [Google Scholar]
- 13.Ito Y, Hirano T. First successful amplification of 18S ribosomal DNA of Cordyceps spp. by the PCR method. Mycoscience. 1996;37:109–110. [Google Scholar]
- 14.Ito Y, Hirano T. The determination of the partial 18 S ribosomal DNA sequences of Cordyceps species. Lett Appl Microbiol. 1997;25:239–242. doi: 10.1046/j.1472-765x.1997.00203.x. [DOI] [PubMed] [Google Scholar]
- 15.Liu Z-Y, Yao Y-J, Liang ZQ, et al. Molecular evidence for the anamorph-teleomorph connection in Cordyceps sinensis. Mycol Res. 2001;105:827–832. [Google Scholar]
- 16.O'Donnell K, Nirenberg HI, Aoki T, et al. A multigene phylogeny of the Gibberella fujikuroi species complex: detection of additional phylogenetically distinct species. Mycoscience. 2000;41:61–78. [Google Scholar]
- 17.O'Donnell K, Kistler HC, Tacke BK, et al. Gene genealogies reveal global phylogeographic structure and reproductive isolation among lineages of Fusarium graminearum, the fungus causing wheat scab. Proc Natl Acad Sci USA. 2000;97:7905–7910. doi: 10.1073/pnas.130193297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dettman JR, Jacobson DJ, Taylor JW. A multilocus genealogical approach to phylogenetic species recognition in the model eukaryote Neurospora. Evolution. 2003;57:2703–2720. doi: 10.1111/j.0014-3820.2003.tb01514.x. [DOI] [PubMed] [Google Scholar]
- 19.Ko KS, Joung HS. Three nonorthologous ITS1 types are present in a polypore fungus Trichaptum abietinum. Mol Phylogenet Evol. 2002;23:112–122. doi: 10.1016/S1055-7903(02)00009-X. [DOI] [PubMed] [Google Scholar]
- 20.Baldwin BG, Sanderson MJ, Porter JM, et al. The ITS region of nuclear ribosomal DNA: A valuable source of evidence on angiosperm phylogeny. Ann Mo Bot Gard. 1995;82:247–277. [Google Scholar]
- 21.Vogler AP, DeSalle R. Evolution and phylogenetic information content of the ITS-1 region in the tiger beetle Cicindela dorsalis. Mol Biol Evol. 1994;11:393–405. doi: 10.1093/oxfordjournals.molbev.a040121. [DOI] [PubMed] [Google Scholar]
- 22.Zijlstra C, Lever AEM, Uenk BJ, et al. Differences between ITS regions of isolates of root-knot nematodes Meloidogyne hapla and M. chitwoodi. Phytopathology. 1995;85:1231–1237. [Google Scholar]
- 23.Buckler ES, Ippolito A, Holtsford TP. The evolution of ribosomal DNA: divergent paralogues and phylogenetic implications. Genetics. 1997;145:821–832. doi: 10.1093/genetics/145.3.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Birdsell JA. Integrating genomics, bioinformatics, and classical genetics to study the effects of recombination on genome evolution. Mol Biol Evol. 2002;19:1181–1197. doi: 10.1093/oxfordjournals.molbev.a004176. [DOI] [PubMed] [Google Scholar]
- 25.Rocha EPC, Danchin A. Base composition bias might result from competition for metabolic resources. Trends Genet. 2002;18:291–294. doi: 10.1016/S0168-9525(02)02690-2. [DOI] [PubMed] [Google Scholar]