Abstract
During myocardial infarction, myocytes die and are replaced by a specialized fibrotic extracellular matrix, otherwise known as scarring. Fibrotic scarring presents a tremendous hemodynamic burden on the heart, as it creates a stiff substrate, which resists diastolic filling. Fibrotic mechanisms result in permanent scarring which often leads to hypertrophy, arrhythmias, and a rapid progression to failure. Despite the deep understanding of fibrosis in other tissues, acquired through previous investigations, the mechanisms of cardiac fibrosis remain unclear. Recent studies suggest that biochemical cues as well as mechanical cues regulate cells in myocardium. However, the steps in myofibroblast transdifferentiation, as well as the molecular mechanisms of such transdifferentiation in vivo, are poorly understood. This review is focused on defining myofibroblast physiology, scar mechanics, and examining current findings of myofibroblast regulation by mechanical stress, stiffness, and topography for understanding fibrotic disease dynamics.
Keywords: biomechanics, cardiac engineering, fibrosis, mechanoregulation, myofibroblasts
1. Introduction
In the United States and many other nations, cardiac failure is the leading cause of death. Heart diseases account for over 800 000 deaths per year (1 in every 3 deaths), and economic expenses exceed $320 billion in direct and indirect costs.[1] Nearly all forms of cardiovascular disease are associated with myocardial fibrosis, which is primarily mediated by cardiac fibroblasts. While cardiac fibroblasts are responsible for extracellular matrix (ECM) maintenance in healthy myocardium, they can also transform into myofibroblasts. Post-transformation, they can contribute to the secretion of cytokines, deposition of ECM, structural support, and filling the mechanical load created by myocyte necrosis.[2] Myofibroblast transdifferentiation is essential in overcoming cardiac injury, but progressive fibrosis often leads to remodeling of both infarcted and residual noninfarcted myocardium. This remodeling results in reduced tissue compliance, increased matrix stiffness, irregular action potential propagation, and progressive heart failure[2] (Figure 1). The limited regenerative capacity of the mammalian myocardium intensifies the fibrotic and inflammatory response during cardiac wound healing.[3–5] These changes lead to disruption of overall tissue organization, critically damaging organ function through hypertrophy, chamber dilation, biochemical intra-cellular signaling factor secretion, and transdifferentiation of neighboring fibroblasts. Fibrosis is linked to ventricular arrhythmias, hypertension, diabetes, rheumatic heart diseases, hypertrophic cardiomyopathy, heart failure, and sudden cardiac death.[6,7] Currently, clinical strategies to combat damage to the myocardium from fibrosis are essentially palliative. This is especially true due to the limited supply of hearts for transplantation[2] and lack of understanding of the regulation of the remodeled cardiac environment on scar tissue formation.
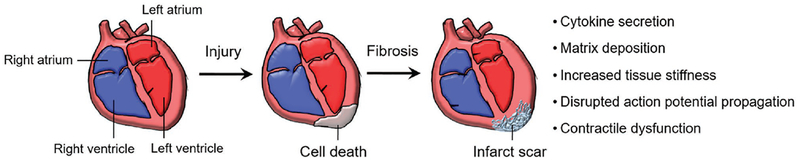
Figure 1.
Illustration of cardiac fibrosis development. Cardiac infarction often results in myocyte necrosis, which is replaced by infarct scar. Cytokine secretion, matrix deposition, increased tissue stiffness, disrupted action potential propagation, and contractile dysfunction are consequential events postinfarction and may lead to heart failure.
Inhibition of heart scarring and fibrosis would be an ideal therapeutic strategy for treating heart diseases. One such disease is Ischemic fibrosis, where obstruction of the coronary arteries leads to a reduction of the oxygen supply to the myocardium. This can potentially result in infarction, where the lack of oxygen results in necrosis. The area of necrosis will eventually be replaced by fibrotic scar, greatly affecting the functionality of the myocardium.
Myocardium is a complex, highly ordered system, with a mix of cellular and acellular components, providing resident cells with strong structural organization as a whole.[2–4] Ultrastructural analysis of mammalian myocardial tissue highlights that the arrangement of aligned cells correlates strongly with the direction of the underlying ECM fibers.[5] In the myocardium, the ECM is aligned congruently, providing a natural direction for myocyte exertion of contractile forces and a defined axis for action potential propagation. In a healthy myocardium, the fibrous ECM provides several other functions as well. These include providing a native myofascial plane between layers of muscle, a barrier to electrical activation of the atria and ventricles, and structural guidance to blood vessels. These functions are often disrupted after myocardial infarction.[4] Postinfarction remodeling, such as ECM deposition, increased stiffness, and impaired contraction, is known to be regulated by chemical, mechanical, and structure cues through myofibroblast transdifferentiation.[8] This suggests that ECM offers mechanical cues for cardiac cellular and macroscopic tissue organization and development.
There is a tremendous need for the development of effective interventions for cardiac fibrosis both clinically and economically. Currently, numerous publications have been made regarding myofibroblasts and cardiac wound healing. Recent publications have shown cell–matrix regulation to play a key role in cardiac wound healing. This review will focus on the current status of research revolving around the mechanical regulation of myofibroblasts in cardiac fibrosis and wound healing, as well as the future targets for possible therapeutic development.
2. Cellular Origins of Cardiac Fibrosis
Myofibroblasts are responsible for secreting a fibrotic matrix in response to injury signals. They can secrete large amounts of matrix proteins including collagen type I, collagen type III, collagen type IV, periostin, and fibronectin.[9] They play a critical role in wound healing in various organs including the lungs, liver, kidneys,[10,11] skeletal muscle,[12] and heart.[13,14] Their contribution to wound healing includes migration, wound contraction, recruitment of inflammatory cells, and the remodeling/secretion of ECM to provide structural reinforcement[4,15] (Figure 2). Morphologically, myofibroblasts are identified by ruffled membranes, a spindle shaped morphology, dendritic processes, and large endoplasmic reticulum organelles. The characteristics of myofibroblast are a cross between fibroblasts and smooth muscle cells, including the expression of alpha smooth muscle actin (αSMA) and the intermediate filament desmin. The contractile property of myofibroblasts originates from the electron dense smooth muscle myosin and αSMA. While these characteristics are all documented, the steps and molecular mechanisms in myofibroblast transdifferentiation in vivo are not well understood.
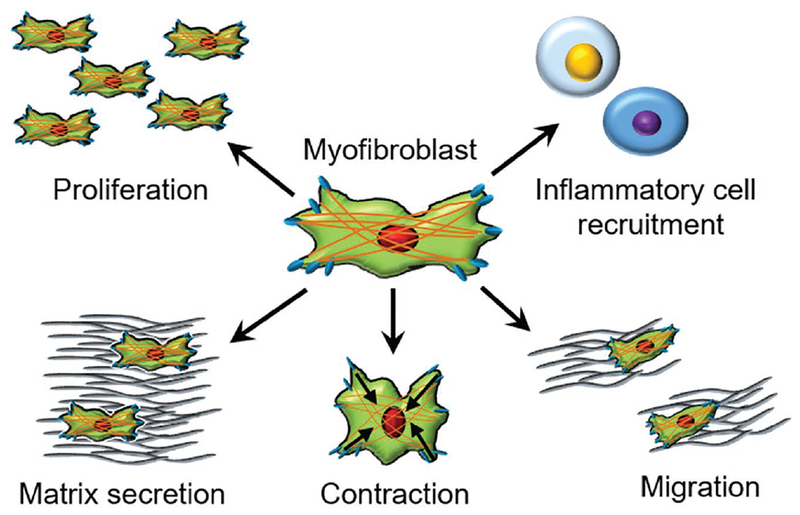
Figure 2.
Myofibroblast characteristics during myocardial fibrosis. Cardiac myofibroblasts are transdifferentiated and proliferate to infarct region. Myofibroblasts are responsible for recruiting inflammatory cells via secretion of proinflammatory factors to the infarct region. Matrix secretion, contraction, and migration of myofibroblast all contribute to compensating tissue load, localization, and wound contraction during cardiac fibrosis.
A few extracellular ligands which are involved in fibroblast to myofibroblast transdifferentiation include transforming growth factor β (TGFβ), endothelin 1,[16] angiotensin-II,[17] nerve growth factor,[18] thrombin,[19] Wnt β catenin/fizz1,[20,21] platelet-derived growth factor (PDGF),[22] and intracellular stress.[23] Previous studies in vitro suggest that in the early stage of transdifferentiation, fibroblasts exhibit an increase in focal adhesion proteins, which increase mechanical stress on the cells.[24,25] As myofibroblast fully differentiates, smooth muscle actin expression increases.[25] Although the presence of αSMA is considered to be a marker for myofibroblasts, longer focal adhesions, paxillin, tensin, ED-A fibronectin, increased αvβ3 or αvβ5 integrin, and excessive secretion of collagen are all collectively used to identify myofibroblasts.[4,26] However, many markers fail to specifically identify cardiac myofibroblasts. This remains a major challenge in cardiac tissue engineering.[27] It should also be noted that multiple factors including inflammatory cytokines such as TGFβ are known to lead fibroblasts to a myofibroblast lineage. However, the factors that initiate and differentiate fibroblasts into myofibroblasts have not been confirmed in vivo.
While normally not present in healthy myocardium, myofibroblasts appear and transform the myocardium upon cardiac injury, in pathological responses, or aging. Importantly, myofibroblasts develop dense microfilaments and actin cytoskeletons that extend the membrane of the cell to an adhesion complex, fibronexus.[28] Altogether, a mature adhesion complex with internal stress fibers generates a contractile force, which is then reinforced by deposition of collagen.[29] Despite this contractile machinery, myofibroblasts are nonexcitable cells that do not directly participate in contractile behavior or conduction of action potentials through gap junctions of normal myocytes. Although Cx43 protein has been reported to be present between fibroblasts and myocytes in the sinus node of a normal rabbit heart, further investigation suggested that such coupling levels are very low, even slowing electrical conduction.[30] Overpopulation of myofibroblasts is likely to hinder myocyte to myocyte coupling and cardiac conduction via gap junctions, while leading to over-stiffening of the myocardium by excessive ECM deposition.[31] Although multiple animal models have shown myofibroblasts play an important role in physiologic remodeling and wound closure, overexpression of the myofibroblasts phenotype often leads to uncontrolled fibrosis.[32–34] In the case of cardiac tissue, this results in pathological ventricular remodeling, hypertrophy, arrhythmia, and even heart failure.
Classes of cardiac fibrosis include reactive interstitial, replacement, and perivascular fibrosis.[35] Reactive interstitial and perivascular fibrosis are often observed in left ventricular pressure overload models or in hearts affected by hypertension, diabetes, or aging.[36,37] Reactive interstitial and perivascular fibrosis are characterized by progressive collagen accumulation in the perivascular and interstitial spaces in the absence of myocyte cell death. Such changes are accompanied by myocyte hypertrophy, and are reported to affect even remote noninfarcted myocardium. Progressive fibrosis is considered a hallmark of aging in many organs including the cardiovascular system. Although aged hearts may exhibit a normal ejection fraction and contractility, the myocardial compliance and ventricular mass are often increased due to deposited collagen from progressive fibrosis.[38] Different types of progressive fibrosis may have different causal mechanisms leading to cardiac fibrosis. Fibrosis induced from hypertension is caused by increased collagen synthesis, but age-induced fibrosis exhibits decreased collagen synthesis, but significant attenuation of matrix-degrading pathways accounting for cumulative collagen deposition.[39] Reactive interstitial and perivascular fibrosis are considered an intermediate marker of fibrosis, as it precedes irreversible replacement fibrosis. Some therapeutic approaches were found to reverse such phenotypes.[40] By contrast, replacement fibrosis is observed in acute myocardial infarction and ischemic heart disease with no effective therapeutic approaches. In this case, necrotic myocytes are replaced with fibrotic scar through excessive matrix deposition, mainly type I collagen.[3] Interstitial and perivascular fibrosis ultimately lead to replacement fibrosis which then often leads to heart failure.
Recent studies suggest that myofibroblasts are primarily derived from resident fibroblasts which undergo programmed transdifferentiation. However, endothelial-derived fibro-blasts,[41] epithelial-derived fibroblasts,[41,42] circulating fibrocytes,[43] perivascular cells,[26] and mesenchymal cells from the Gli1 lineage may also contribute to the population of myofibroblasts within injured tissue.[4] The diversity of myofibroblast precursor cells is one of the confounding factors in understanding myofibroblast function, as well as their role in fibrotic remodeling of the heart after injury or during disease progression.
3. Microenvironmental Cardiac Scar Mechanics
Due to the recent findings of mechanical cues modulating myofibroblast transdifferentiation,[25,44–47] cardiac researchers are focusing on investigating the signaling pathways underlying the transduction of mechanical cues. Traditional understandings of the myofibroblast and its role in cardiac wound healing have mainly relied on an in vitro setting on flat tissue culture plastic. These settings are distant from the rich in vivo microenvironments, which myofibroblasts continuously interact with both before and after transdifferentiation.
In vivo, fibroblasts and myofibroblasts regulate the ECM dynamically, and concurrently receive environmental regulatory cues. The heart’s ECM microenvironment is known to maintain the heart’s electrophysiology, provide structural support to myocytes, and provide residing cells with signaling proteins.[3] Studies on ECM dynamics have found that ECM not only regulates fibrosis chemically, but also mechanically.[8,45,48,49] The conversion of mechanical signals into biochemical signals plays a pivotal role in cellular differentiation. A large range of subcellular structures have been found to contribute to this process. Some of these include the ECM itself, cytoskeletal filaments, myosin motors, growth factors, integrins, and stretch activated ion channels.
The healthy heart is organized with various ECM proteins, which contribute to synchronized contraction, tight cell–cell coupling, and directional action potential propagation.[48,50] Since directional ECM organization has been observed in many studies, cardiac tissue engineering has been attempting to recapitulate the native heart ECM to understand the mechanisms of cell–ECM interactions.[8,48,50–52] A major protein in the ECM of the heart is planar laminin. It is reported that healthy adult myocardium is comprised of about 35% laminin, however as an infarct develops, collagen, specifically type I collagen, a fibrillary protein that provides tensile strength and stiffness to myocardium, dominates[53] (Figure 3). The increase in fibrillar type I collagen content, resulting in increased rigidity, is generally a hallmark of fibrosis.[54] Both collagen content and tissue stiffness in the microenvironment are known to be regulated by myofibroblasts. Fibrillar collagen, which is often absent in healthy myocardium, develops on the border of an infarct in congruent direction to myocytes upon injury.[55] Notably, the core of an infarct displays random orientation of collagen fibers (Figure 4). The mechanoregulation of remodeled microenvironments in infarct scaring has not been investigated in depth. Cyclic stretch,[56–58] rigidity,[49,56] ECM orientation,[59] infarct location,[45] and topographic cues[48,50] are beginning to be examined for roles in cardiac scar formation and myofibroblast regulation. Studies suggest these environmental mechanical cues are key regulators in cardiac remodeling. Attempts made to recapitulate in vivo microenvironments, in order to investigate effects of exogenous mechanical cues, would advance effective study of cellular biomechanical function.
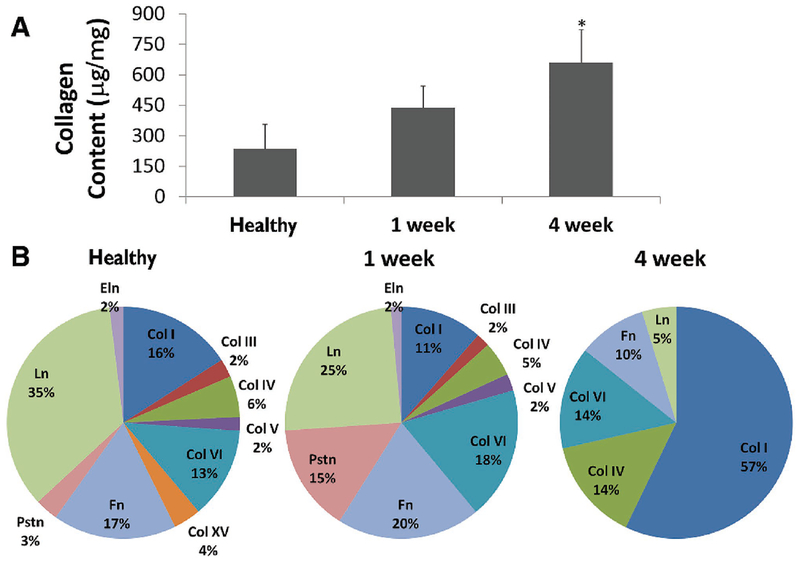
Figure 3.
Extracellular matrix composition is remodeled postinfarction. A) Total collagen content within the 4-week infarct is significantly greater than the both healthy and 1-week conditions. B) Liquid chromatography mass spectrometry coupled mass spectrometry (LC-MS/MS) spectrum count analysis has shown the relative percentages of each matrix protein content within the decellularized heart. Note that Pstn is periostin, Ln is laminin, Eln is elastin, Fn is fibronectin, and Col is collagen. Reproduced with permission.[53] Copyright 2014, BioMed Central Ltd.
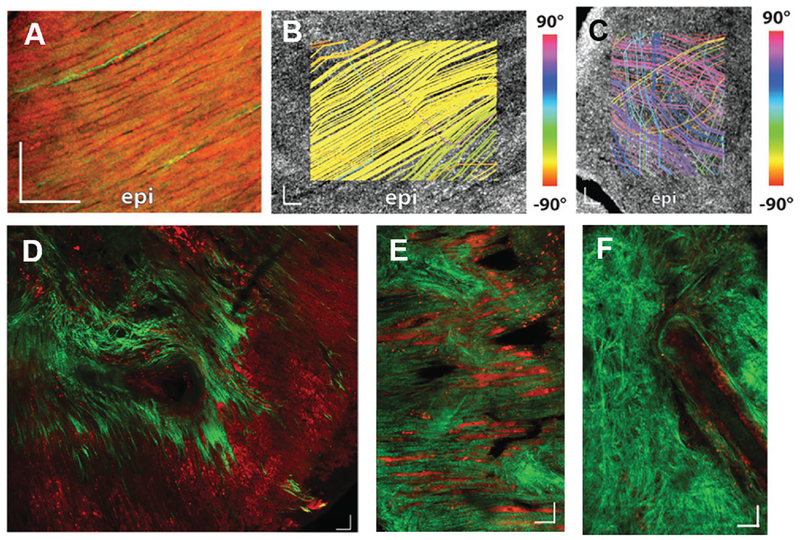
Figure 4.
Myofiber and collagen direction is remodeled during cardiac fibrosis. Myofiber direction was analyzed for A,B) healthy and C) infarcted heart. A) Two-photo microscopy and tractography streamline analysis of B) healthy and C) infarcted heart displayed a significant disruption of myofiber direction on infarcted heart. D) Collagen (green) direction was regionally remodeled on the infarct site. E) Collagen fibers were congruently aligned to cardiomyocytes (red) on the border of the infarct. F) Core of infarct exhibited randomly oriented collagen fibers. Reproduced under the terms of the CC-BY license.[55] Copyright 2016, The Authors.
4. Mechanical Stress Underlies Fibroblast to Myofibroblast Differentiation
Myofibroblasts are highly sensitive to mechanical force, and can generate contractile tension on their surroundings during wound healing. Mechanical forces are known to induce increased proliferation, reduced collagenolytic activity, and increased collagen production.[60] A number of studies are starting to report congruent effects of mechanical and chemical signals on myofibroblast transdifferentiation.[9,61,62] Evidences suggest cyclic tension itself (15%, 1 Hz) can transdifferentiate fibroblasts into myofibroblasts, without any secondary soluble growth factors.[56–58,60] On the contrary, a number of studies also report that cyclic strain has an inhibitory effect on myofibroblast transdifferentiation.[63–66] Here, the inhibitory effect was reverted with increased anisotropy, suggesting that fibroblasts can also sense the directionality of cyclic strain.[63] Moreover, infarcts induced on the equator of hearts, where myocytes contract in a circumferential direction, change the orientation of ECM fibers. This was not observed when the infarct was restricted to the apex, where myocytes induce circumferential and longitudinal contraction of the heart.[45]
Integrins are well-established mechanosensors in fibroblasts and myofibroblasts that connect the ECM and cytoplasmic actin cytoskeleton. These mechanosensors are heterodimer membrane receptor proteins. They are composed of an alpha and a beta chain which confer specificity to certain ECM components. Integrins link the actin/myosin cytoskeleton within fibroblasts to the ECM, allowing cells to exert and sense mechanical forces in the external environment. These mechanosensors have been shown to play a role in myofibroblast transdifferentiation.
Myofibroblast differentiation and cell specific markers have been shown to increase with mechanical tension.[60,67] Such increased transdifferentiation events were effectively reduced with the inhibition of integrins, specifically integrin αv, in liver, kidneys, and lungs via suppression of latent TGFβ activation.[68] Stretch-mediated mechanical signals have also been shown to alter ECM–integrin interactions and vary cellular responses.[69] Thus, strategies to modulate myofibroblast integrin inhibition with small molecules or antibodies are emerging as a novel method of combating fibrotic cardiac diseases.[70]
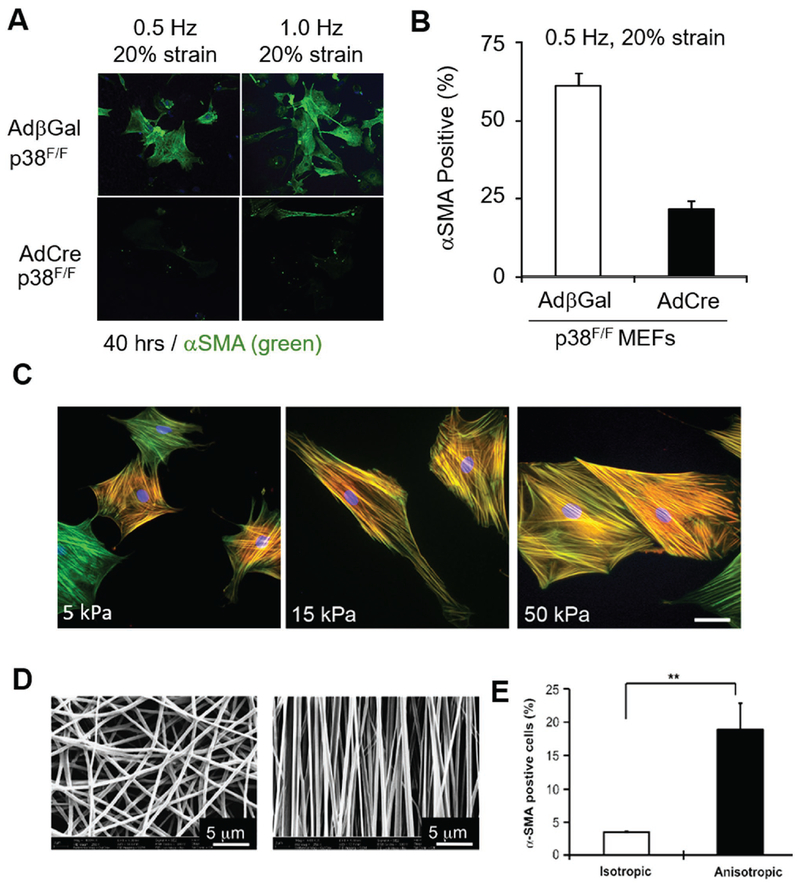
One significant pathway that transduces mechanical stress is the mitogen-activated protein kinases (MAPK). All three MAPK kinases (ERK, JNK, and p38) were activated with cell stretch, and inactivated with cell contraction.[71] A number of results suggest there exists a selective activation pathway of MAPK during mechanical force stimulation.[60,72,73] Interestingly, passive biaxial stretching has shown an increase in ERK and JNK activity, but not the p38 kinase pathway.[74,75] Conversely, tensile forces were shown to increase activity of p38, but not ERK and JNK.[76] Recent findings have shown that increases in myofibroblast transdifferentiation induced by cyclic strain were inhibited with p38 knockout, suggesting the regulatory effect of p38 on transducing mechanical cues for fibrotic response[64] (Figure 5A,B).
Figure 5.
Myofibroblast transdifferentiation is mechanically regulated. A,B) Cyclic strain has shown regulatory effect on myofibroblast transdifferentiation. Recent findings suggest p38 plays a key role in mechanoregulation of cyclic strain. Reproduced with permission.[64] Copyright 2017, Lippincott Williams & Wilkins. C) Stiffness has shown to regulate fibroblast transdifferentiate into myofibroblast. An increase in elastic modulus has shown increased expression of αSMA. Reproduced with permission.[85] Copyright 2013, Public Library of Science. D) Myofibroblasts are also sensitive to alignment cues imposed by electrospun fibers in isotropic (left) and anisotropic (right) fibers. E) Fibroblasts cultured on anisotropic fibers have expressed higher αSMA signals. D,E) Reproduced with permission.[100] Copyright 2012, Elsevier.
Unlike myocytes, myofibroblasts do not produce cyclic tension. However, studies suggest that static tensile forces can also regulate myofibroblast fate. A static tensile force of 0.65 pN mm−2 resulted in a twofold increase in αSMA protein levels within a short period for low basal levels of αSMA. While a decrease in αSMA for high basal levels of αSMA through MAPK occurs.[60] It was found that stress worked synergistically with TGFβ, causing activation of latent TGFβ1, which in turn induces myofibroblast differentiation.[46,77] Activation of TGFβ1 by release from the latency associated peptide has been found to require as low as 40 pN of integrin-transmitted force.[77] Mechanical force induces myofibroblast differentiation, setting up a positive feedback loop, in which newly differentiated myofibroblasts exert force on the surrounding microenvironment. This positive feedback is assisted by TGFβ1 activation, amplifying the inductive signals for a fibroblast to differentiate.
Incorporation of αSMA into stress fibers is significant, as it leads to myofibroblast contractility. The contractile stress fibers, comprised of mature actin microfilaments and non-muscle myosin are regulated by myosin light-chain (MLC) phosphorylation.[8] This phosphorylation is regulated by Rho kinase. RhoA, a small GTPase protein of the Rho family, is known to enhance actin reorganization and activate TGFβ responses. This kinase is another major factor in myofibroblast transdifferentiation.[78,79] Inhibition of RhoA significantly reduces contractile force, as well as wound granulation in tissue contraction.[80]
RhoA/Rho-associated kinase (ROCK) regulates not only the phosphorylation status of MLC, but also underlines the remodeling of the actin cytoskeleton into stable stress fibers.[81] Although both ROCK1 and ROCK2 have been implicated in cardiac hypertrophy and ventricular remodeling, ROCK1 is central to the development of cardiac fibrosis. In the context of mechanical signaling the ROCK kinases are critical for mechanosensing in both fibroblasts and myofibroblasts.[81] The close relationship between external mechanical tensions, myofibroblast induced tension, the pathways involved, and fate changes all suggest that focus on these areas may represent a new strategy in preventing maladaptive scar formation and in developing an antifibrotic therapeutic strategy.[82]
5. Substrate Stiffness Regulates Myofibroblast Fate
Recent findings demonstrate that myofibroblast fate is also regulated by substrate stiffness.[83–85] Fibroblasts cultured on low stiffness planar substrates do not form stress fibers or express αSMA, in contrast to fibroblasts cultured on high stiffness substrate or tissue culture plastic[85,86] (Figure 5C). It has been discovered that tissue stiffness or loading has a close relationship to wound healing and scar formation. However, the understanding of the molecular mechanism of how myofibroblasts transduce mechanical elasticity is still at an elementary stage. Various models have been suggested to understand how myofibroblasts sense external stiffness. One model is detection through mechanosensitive membrane ion channels, which change conformation in response to external tension.
Mechanosensitive ion channels were shown to detect rapid changes in tension induced by magnetic beads.[87] Mechanosensitive ion channel models are appealing, however they have drawbacks. This model does not reflect in vivo fibrotic response, as matrix dynamics occur over a period of weeks to even months, while the ion channel models are limited to short term changes. Thus, in order to understand the process of myofibroblast fate change mediated by substrate stiffness, the current focus is on investigating ECM–cell adhesion dynamics and cytoskeletal regulation through mechanotransduction.
Formation of enlarged focal adhesions has been found to be a key step in a feedback loop of external stiffness to actin stress fiber organization that regulates myofibroblast transdifferentiation. Fibronexus, a myofibroblast-specific extensive ECM–cell adhesion complex, is formed in fibrotic tissue by myofibroblasts.[4] In vitro, enlarged focal adhesion complexes (“supermature focal adhesion”) are often observed in high stiffness substrates. Limiting myofibroblasts focal adhesion formation on arrays of restricted islets led to the rapid loss of αSMA expression.[88] Moreover, increasing the size of the islets to allow supermature focal adhesion formation on extendable membranes reincorporated αSMA regardless of the stretch variable.[88] However, it is not clear whether formation of supermature focal adhesions is the primary event which determines myofibroblast fate post injury.
Approaches using 3D model systems with loaded ECM may provide the most relevant environment for simulating in vivo interaction between myofibroblasts and ECM.[88] However, the unrestrained ECM gels with loaded fibroblasts do not provide a continuous positive feedback of increased stiffness and myofibroblast interaction.[89] When collagen gel loses its elasticity, myofibroblasts lose their stress fibers and fibronexus adhesion complexes, which adhere myofibroblasts to collagen fibrils.[90] This is similar to the case when myofibroblasts cultured on stiff substrates are treated with actin-myosin inhibitors.[28] Thus, it is believed that substrate stiffness dynamically and continuously regulates myofibroblast transdifferentiation.
This hypothesis of substrate stiffness regulating myofibroblasts is backed by recent findings which indicate Yes-associated protein (YAP) and transcriptional coactivator with proteins Psd-95, DlgA, and ZO1 (PDZ) binding domain (TAZ), which are transcriptional coactivators of the Hippo pathway, may be closely involved in myofibroblast transdifferentiation in response to ECM stiffness.[47] Where YAP and TAZ are not expressed in healthy tissue, they have been found to be expressed in fibrotic tissue. YAP and TAZ are unique in a way that their responses are mediated by nuclear translocation of the YAP/TAZ complex followed by altered gene expression.[91,92] YAP and TAZ have been found to regulate various cell fates in response to mechanical cues.[91,92] In stiff substrate cultures in vitro, YAP and TAZ accumulate in the nuclei of fibroblasts. This accumulation results in profibrotic matrix synthesis, contraction, and proliferation. This response is through plasminogen activator inhibitor (PA-1) regulation, independent of TGFβ signaling.[47] Knockdown of YAP and TAZ has been found to suppress the myofibroblastic response. Utilization of a “smart polymer,” with the ability to change stiffness has also revealed that a switch in substrate stiffness regulates YAP and TAZ activation.[91] Although the role of YAP and TAZ in cardiac fibrosis has not been investigated in depth, YAP activation was shown to increase cardiac function and enhanced regeneration.[93,94]
Not only does mechanical stiffness regulate enhanced trans-differentiation of fibroblasts, it also synergistically modulates various stimulus induced responses. It is reported that matrix stiffness modulates TGFβ induced transdifferentiation, with a significantly higher response to TGFβ on stiffer substrates.[83] It has also been found that contraction of myofibroblasts promotes latent TGFβ activation on a stiffened matrix; where the activation of TGFβ via integrin-mediated myofibroblast contraction could be a critical point in fibrosis.[78] Traditionally, mechanical activation of myofibroblast differentiation was perceived as an acute process that is limited to contractile force of cells. However, evidences suggest that even prestressed ECM can mechanically prime late stage transdifferentiation.[46]
It has been found that in response to matrix stiffening, actin dynamics change causing filamentous actin polymerization to be more favorable. This results in nuclear translocation of myocardial related transcription factor (MRTF), a marker for myofibroblast differentiation, which has a part in regulating expression of αSMA.[84] In this case, both actin and MRTF may mediate an intrinsic mechanotransduction pathway, that regulates fibroblasts differentiating to myofibroblasts induced by matrix stiffening.[84] Increased expression of actin in response to matrix stiffness is thought to be sufficient in driving myofibroblasts to generate contractile force.[8] Identified as a mechanosensitive protein, αSMA localizes to stress fibers under external mechanical load. Although mechanisms underlying αSMA dynamics on myofibroblast stiffening are not known, evidence shows intracellular inhibition of αSMA increases motility, and reduces contraction both in vitro and in vivo.[25] Substrate stiffness reduction also leads to disassembly of αSMA from stress fibers, suggesting an interactive nature to stiffness regulation.
6. Matrix Topography Regulates Myofibroblast Fate
The heart is an organ that exhibits exceptionally high anisotropy of both myocytes and fibroblasts. This anisotropy and cell–cell junctions contribute to synchronized electric signal propagation and contraction.[50] Previously, ECM topography and anisotropy have shown to play a critical role in controlling cell and tissue function.[50] Electron microscopy of the myocardium has confirmed a directional ECM underlying cells.[50] Not only does the ECM supply cells with chemical cues to adhere, it also physically provides mechanical structures for cells to bind. While the mechanisms of chemical binding have been studied carefully, the role of ECM and its ability to impart regulatory mechanical cues, through variations such as the dimension of fiber bundles, orientation, and density, have not been thoroughly studied in the field of cardiac fibrosis.
The traditional method of culturing myofibroblasts often utilizes a smooth surface substrate, with uncontrolled ECM organization. The fundamental properties and functions of fibroblasts such as migration and cell fate can be affected by engineering mechanical properties of the culture matrix.[95–98] In this context, mechanically modifying the matrix to establish a physiologically relevant environment is critical to creating cardiac scar tissue models to study cardiac fibrosis.
The native anisotropic morphology is lost when cardiac cells are maintained in vitro using standard cell culture substrates and techniques. Infarcted hearts have shown disrupted matrix organization when compared to healthy hearts which consists of a left handed helix matrix.[55] Moreover, the heart matrix content shifts from sheet-like laminin-dominant condition to a fibrous collagen-dominant condition.[53] This loss of matrix organization disrupts the structural organization of ECM cues with adverse consequences for cardiac cell physiological properties.
Analysis of infarct scar remodeling has shown that matrix directionality is regionally regulated on the border and core of the infarct scar.[55] The core of the infarct consisted of matrix oriented in random directions, while the border of the infarct scar displayed an aligned fibrous matrix. To develop relevant model systems to study fibrosis, several engineering approaches have been attempted to recapitulate the dimensions, directionality, and spacing of native ECM fibers in healthy and scarred tissues.[96,98,99]
Clinically, the location of the infarct has been reported to affect ECM orientation in scar tissue.[45] By contrast, initial infarct size or orientation did not have a significant effect on ECM remodeling. Infarcts induced near the equator of the left ventricle resulted in scar ECM remodeling in a circumferential direction, whereas infarcts induced near the apex resulted in isotropic organization of the ECM.[45] These results suggest that mechanical regulation in infarct scars is a complex system with outcomes related to the location of infarcts. The fibroblast cells exhibited more directional and higher cell migration speeds on anisotropic nanoscale fibers over isotropic nanofibers and 3D models of wounds.[100,101] Moreover, anisotropic cues also regulate fundamental fibroblast cell fate by differentiating to a myofibroblastic lineage[46,100] (Figure 5D,E). Specifically, the integrin β1 signaling pathway was activated, and phosphorylation of focal adhesion kinase was observed in response to anisotropic cues.[100]
It is well known that myofibroblasts are capable of remodeling their surrounding ECM. Recent studies have found that this remodeled ECM mechanically induces a myofibroblastic response.[46] In these studies, myofibroblasts deposited similar levels of latent TGFβ as fibroblasts, while the organization of latent transforming growth factor beta binding protein-1 (LTBP-1) differed. Myofibroblasts were found to organize LTBP-1, an integral component of the ECM that stores and presents latent TGFβ, to denser and straighter fibrils. Pathologically organized ECM’s ability to trigger enhanced latent TGFβ activation may explain how decellularized ECM from fibrotic tissue leads de novo seeded cells to fibrotic properties even in the absence of TGFβ treatment, while decellularized ECM from normal tissue did not.[102]
Although not as thoroughly investigated as other mechanical regulatory cues, directionality of ECM indeed plays a critical role in myofibroblast transdifferentiation. In this context, mechanically modifying the matrix to establish a physiologically relevant environment is essential. It aids in developing a fibrotic scar tissue model for understanding mechanisms underlying myofibroblast fate mapping. This may enable development of new therapeutic approach, targeting ECM directly for alleviating fibrosis.
7. Conclusions
Myofibroblasts are an essential cell type in the heart, heavily involved in both damage repair and maintenance of cardiac function. In this review, we discussed the microenvironmental control of myofibroblast fate and fibrosis (Figure 6). Both chemical and mechanical signaling were highlighted in comparing healthy hearts to damaged hearts. Fibroblasts as well as their transdifferentiated myofibroblast cells have shown sensitivity to various mechanical signals, and quite a few signaling pathways have been proposed to transduce such signals. Previous studies have been conducted to investigate the chemical regulation of fibroblast differentiation, garnering some level of understanding. However, our understanding of the mechanoregulation of myofibroblast transdifferentiation and fibrosis regulation remains limited. What is known is that during fibrosis, heart tissue experiences a number of mechanical changes, and such changes act in synergy with chemical cues to induce various fibrotic responses through various signaling pathways. The complex chemical and mechanical signals that interplay in vivo present great challenges to therapeutic cardiac engineering. Recent transgenic studies show promising results in understanding the origins[103,104] and mechanics[32,33,82,105] of myofibroblast transdifferentiation. Such approaches with mechanically engineered platforms could synergistically contribute to accurate assessment of myofibroblast regulation during fibrosis. Emergence of biomedical micro-electromechanical technology may contribute to the field by developing mechanically engineered constructs, to simulate an accurate microenvironment, allowing for the study of the mechanics behind cardiac fibrosis. Decellularized matrix analysis with biomimetic scaffold designs may enable cell culture systems that could recapitulate microenvironmental cues of the in vivo myocardium, and may provide a faithful model to increase our understanding of the mechanical regulation of cardiac fibrosis. Moreover, 3D systems which offer explicit control over factors such as, cyclic stretch, rigidity, and topographic cues, would be a valuable asset, providing insights to combat cardiac diseases, as most forms are associated with myocardial fibrosis. A patient-derived stem cell culture in conjunction with a mechanically regulated microenvironment may contribute to personalized-therapy development. With a greater understanding of mechanical regulation, inhibition of heart scarring and fibrosis could become a realistic therapeutic strategy for treating heart diseases.
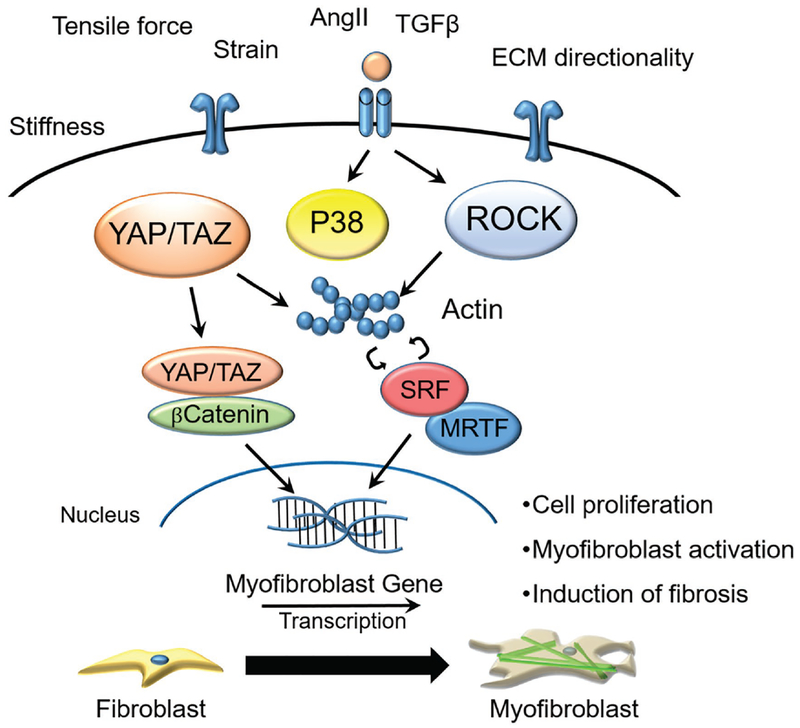
Figure 6.
Schematic illustration of signaling pathways regulating fibroblast transdifferentiation into myofibroblast through mechanoregulation. Various pathways such as YAP/TAZ, p38, ROCK, serum response factor (SRF), and MRTF have been identified to transduce external mechanical cues such as stiffness, tensile force, strain, and ECM directionality.
Acknowledgements
This work was supported by National Institutes of Health grants R21 EB020132 and R01 HL135143 (to D.-H.K.). This work was also supported with funds from the National Institute of Health R00 HL119353 (to J.D.).
Biography

Jennifer Davis is an assistant professor at the University of Washington in the Departments of Pathology and Bioengineering. The Davis lab studies the basic tenets of fibroblast biology and genetically controls fibroblast function to understand the role of the matrix in cardiac biology.

Deok-Ho Kim received his Ph.D. degree in biomedical engineering from The Johns Hopkins University School of Medicine in 2010. He is currently an associate professor in the Department of Bioengineering at the University of Washington. Earlier in his career, he worked at the Swiss Federal Institute of Technology at Zurich (ETH Zürich) and the Korea Institute of Science and Technology as a research scientist. His research interests center on cell-matrix mechanobiology, bioinspired materials/devices, and stem cell and tissue engineering.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Contributor Information
Peter Kim, Department of Bioengineering, University of Washington, Seattle, WA 98109, USA,.
Nick Chu, Department of Bioengineering, University of Washington, Seattle, WA 98109, USA,.
Jennifer Davis, Department of Bioengineering, University of Washington, Seattle, WA 98109, USA,; Center for Cardiovascular Biology, University of Washington, Seattle, WA 98109, USA Department of Pathology, University of Washington School of Medicine, Seattle, WA 98109, USA.
Deok-Ho Kim, Department of Bioengineering, University of Washington, Seattle, WA 98109, USA,; Center for Cardiovascular Biology, University of Washington, Seattle, WA 98109, USA Institute of Stem Cell and Regenerative Medicine, University of Washington, Seattle, WA 98109, USA.
References
- [1].Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, Das SR, Ferranti SD, Despres JP, Fullerton HJ, Howard VJ, Huffman MD, Isasi CR, Jimenez MC, Judd SE, Kissela BM, Lichtman JH, Lisabeth LD, Kiu S, Mackey RH, Magid DJ, McGuire DK, Mohler ER, Moy C, Muntner P, Mussolino ME, Nasir K, Neumar RW, Nichol G, Palaniappan L, Pandey DK, Reeves MJ, Rodriguez CH, Rosamond W, Sorlie PD, Stein J, Towfighi A, Turan TN, Virani SS, Woo D, Yeh RW, Turner MB, Circulation 2016, 133, e38. [DOI] [PubMed] [Google Scholar]
- [2].Segers VF, Lee RT, Nature 2008, 451, 937. [DOI] [PubMed] [Google Scholar]
- [3].Spinale FG, Physiol. Rev 2007, 87, 1285. [DOI] [PubMed] [Google Scholar]
- [4].Hinz B, J. Invest. Dermatol 2007, 127, 526. [DOI] [PubMed] [Google Scholar]
- [5].Song K, Nam YJ, Luo X, Qi X, Tan W, Huang GN, Acharya A, Smith CL, Tallquist MD, Nelson EG, Hill JA, Bassel-Duby R, Olson EN, Nature 2012, 485, 599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Pfeffer JM, Pfeffer MA, Fletcher PJ, Braunwald E, Am. J. Physiol 1991, 260, H1406. [DOI] [PubMed] [Google Scholar]
- [7].Gaudron P, Kugler I, Hu K, Bauer W, Eilles C, Ertl G, J. Am. Coll. Cardiol 2001, 38, 33. [DOI] [PubMed] [Google Scholar]
- [8].Rouillard AD, Holmes JW, J. Physiol 2012, 590, 4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Stempien-Otero A, Kim DH, Davis J, J. Mol. Cell. Cardiol 2016, 97, 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Meran S, Steadman R, Int. J. Exp. Pathol 2011, 92, 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Grande MT, Lopez-Novoa JM, Nat. Rev. Nephrol 2009, 5, 319. [DOI] [PubMed] [Google Scholar]
- [12].Serrano AL, Mann CJ, Vidal B, Ardite E, Perdiguero E, Munoz-Canoves P, Curr. Top. Dev. Biol 2011, 96, 167. [DOI] [PubMed] [Google Scholar]
- [13].van den Borne SW, Diez J, Blankesteijn WM, Verjans J, Hofstra L, Narula J, Nat. Rev. Cardiol 2010, 7, 30. [DOI] [PubMed] [Google Scholar]
- [14].Brown RD, Ambler SK, Mitchell MD, Long CS, Annu. Rev. Pharmacol. Toxicol 2005, 45, 657. [DOI] [PubMed] [Google Scholar]
- [15].Suzuki J, Isobe M, Aikawa M, Kawauchi M, Shiojima I, Kobayashi N, Tojo A, Suzuki T, Kimura K, Nishikawa T, Sakai T, Sekiguchi M, Yazaki Y, Nagai R, Circulation 1996, 94, 1118. [DOI] [PubMed] [Google Scholar]
- [16].Martin P, Tzanidis A, Stein-Oakley A, Krum H, J. Cardiovasc. Pharmacol 2000, 36, S367. [DOI] [PubMed] [Google Scholar]
- [17].Swaney JS, Roth DM, Olson ER, Naugle JE, Meszaros JG, Insel PA, Proc. Natl. Acad. Sci. USA 2005, 102, 437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Micera A, Vigneti E, Pickholtz D, Reich R, Pappo O, Bonini S, Maquart FX, Aloe L, Levi-Schaffer F, Proc. Natl. Acad. Sci. USA 2001, 98, 6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Bogatkevich GS, Tourkina E, Abrams CS, Harley RA, Silver RM, Ludwicka-Bradley A, Am. J. Physiol 2003, 285, L334. [DOI] [PubMed] [Google Scholar]
- [20].Liu T, Dhanasekaran SM, Jin H, Hu B, Tomlins SA, Chinnaiyan AM, Phan SH, Am. J. Pathol 2004, 164, 1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Daskalopoulos EP, Hermans KC, Janssen BJ, Matthijs Blankesteijn W, Trends Cardiovasc. Med 2013, 23, 121. [DOI] [PubMed] [Google Scholar]
- [22].Kong P, Christia P, Frangogiannis NG, Cell. Mol. Life Sci 2014, 71, 549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Shephard P, Hinz B, Smola-Hess S, Meister JJ, Krieg T, Smola H, Thromb. Haemostasis 2004, 92, 262. [DOI] [PubMed] [Google Scholar]
- [24].Santiago JJ, Dangerfield AL, Rattan SG, Bathe KL, Cunnington RH, Raizman JE, Bedosky KM, Freed DH, Kardami E, Dixon IM, Dev. Dyn 2010, 239, 1573. [DOI] [PubMed] [Google Scholar]
- [25].Tomasek JJ, Gabbiani G, Hinz B, Chaponnier C, Brown RA, Nat. Rev. Mol. Cell Biol 2002, 3, 349. [DOI] [PubMed] [Google Scholar]
- [26].Hinz B, J. Biomech 2010, 43, 146. [DOI] [PubMed] [Google Scholar]
- [27].Moore-Morris T, Guimaraes-Camboa N, Banerjee I, Zambon AC, Kisseleva T, Velayoudon A, Stallcup WB, Gu Y, Dalton ND, Cedenilla M, Gomez-Amaro R, Zhou B, Brenner DA, Peterson KL, Chen J, Evans SM, J. Clin. Invest 2014, 124, 2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Chrzanowska-Wodnicka M, Burridge K, J. Cell Biol. 1996, 133, 1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Gabbiani G, J. Pathol. 2003, 200, 500. [DOI] [PubMed] [Google Scholar]
- [30].Thompson SA, Copeland CR, Reich DH, Tung L, Circulation 2011, 123, 2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Thompson SA, Blazeski A, Copeland CR, Cohen DM, Chen CS, Reich DM, Tung L, J. Mol. Cell. Cardiol 2014, 68, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Davis J, Burr AR, Davis GF, Birnbaumer L, Molkentin JD, Dev. Cell 2012, 23, 705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Davis J, Salomonis N, Ghearing N, Lin SC, Kwong JQ, Mohan A, Swanson MS, Molkentin JD, Nat. Commun 2015, 6, 10084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Small EM, J. Cardiovasc. Transl. Res 2012, 5, 794. [DOI] [PubMed] [Google Scholar]
- [35].Anderson KR, Sutton MG, Lie JT, J. Pathol 1979, 128, 79. [DOI] [PubMed] [Google Scholar]
- [36].Wang X, Guo Z, Ding Z, Khaidakov M, Lin J, Xu Z, Sharma SG, Jiwani S, Mehta JL, J. Mol. Cell. Cardiol 2015, 80, 101. [DOI] [PubMed] [Google Scholar]
- [37].Russo I, Frangogiannis NG, J. Mol. Cell. Cardiol 2016, 90, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Upadhya B, Kitzman DW, Heart Failure Clinics 2017, 13, 485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Robert V, Besse S, Sabri A, Silvestre JS, Assayag P, Nguyen VT, Swynghedauw B, Delcayre C, Lab. Invest 1997, 76, 729. [PubMed] [Google Scholar]
- [40].Lopez B, Gonzalez A, Beaumont J, Querejeta R, Larman M, Diez J, J. Am. Coll. Cardiol 2007, 50, 859. [DOI] [PubMed] [Google Scholar]
- [41].von Gise A, Pu WT, Circ. Res 2012, 110, 1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Zeisberg EM, Tarnavski O, Zeisberg M, Dorfman AL, McMullen JR, Gustafsson E, Chandraker A, Yuan X, Pu WT, Roberts AB, Neilson EG, Sayegh MH, Izumo S, Kalluri R, Nat. Med 2007, 13, 952. [DOI] [PubMed] [Google Scholar]
- [43].Souders CA, Bowers SL, Baudino TA, Circ. Res 2009, 105, 1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Carver W, Goldsmith EC, BioMed Res. Int 2013, 2013, 101979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Fomovsky GM, Rouillard AD, Holmes JW, J. Mol. Cell. Cardiol 2012, 52, 1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Klingberg F, Chow ML, Koehler A, Boo S, Buscemi L, Quinn TM, Costell M, Alman BA, Genot E, Hinz B, J. Cell Biol 2014, 207, 283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Liu F, Lagares D, Choi KM, Stopfer L, Marinkovic A, Vrbanac V, Probst CK, Hiemer SE, Sisson TH, Horowitz JC, Rosas IO, Fredenburgh LE, Feghali-Bostwick C, Varelas X, Tager AM, Tschumperlin DJ, Am. J. Physiol 2015, 308, L344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Zong X, Bien H, Chung CY, Yin L, Fang D, Hsiao BS, Chu B, Entcheva E, Biomaterials 2005, 26, 5330. [DOI] [PubMed] [Google Scholar]
- [49].Engler AJ, Carag-Krieger C, Johnson CP, Raab M, Tang HY, Speicher DW, Sanger JW, SAnger JM, Discher DE, J. Cell Sci 2008, 121, 3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Kim DH, Lipke EA, Kim P, Cheong R, Thompson S, Delannoy M, Suh KY, Tung L, Levchenko A, Proc. Natl. Acad. Sci. USA 2010, 107, 565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].van Spreeuwel AC, Bax NA, Bastiaens AJ, Foolen J, Loerakker S, Borochin M, Van der Schaft DW, Chen CS, Baaijens FP, Bouten CV, Integr. Biol 2014, 6, 422. [DOI] [PubMed] [Google Scholar]
- [52].Ott HC, Matthiesen TS, Goh SK, Black LD, Kren SM, Netoff TI, Taylor DA, Nat. Med 2008, 14, 213. [DOI] [PubMed] [Google Scholar]
- [53].Sullivan KE, Quinn KP, Tang KM, Georgakoudi I, Black LD 3rd, Stem Cell Res. Ther 2014, 5, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Weber KT, J. Am. Coll. Cardiol 1989, 13, 1637. [DOI] [PubMed] [Google Scholar]
- [55].Goergen CJ, Chen HH, Sakadzic S, Srinivasan VJ, Sosnovik DE, Physiol. Rep 2016, 18, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Pang Y, Wang X, Lee D, Greisler HP, Biomaterials 2011, 32, 3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Tangkijvanich P, Santiskulvong C, Melton AC, Rozengurt E, Yee HF Jr., J. Cell. Physiol 2002, 191, 351. [DOI] [PubMed] [Google Scholar]
- [58].Wang JH, Yang G, Li Z, Shen W, J. Biomech 2004, 37, 573. [DOI] [PubMed] [Google Scholar]
- [59].Dickinson RB, Guido S, Tranquillo RT, Ann. Biomed. Eng 1994, 22, 342. [DOI] [PubMed] [Google Scholar]
- [60].Wang J, Chen H, Seth A, McCulloch CA, Am. j. Physiol 2003, 285, H1871. [DOI] [PubMed] [Google Scholar]
- [61].Grinnell F, Trends Cell Biol 2000, 10, 362. [DOI] [PubMed] [Google Scholar]
- [62].Numaga-Tomita T, Kitajima N, Kuroda T, Nishimura A, Miyano K, Yasuda S, Kuwahara K, Sato Y, Ide T, Birnbaumer L, Sumimoto H, Mori Y, Nishida M, Sci. Rep 2016, 6, 39383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Gould RA, Chin K, Santisakultarm TP, Dropkin A, Richards JM, Schaffer CB, Butcher JT, Acta Biomater 2012, 8, 1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Molkentin JD, Bugg D, Ghearing N, Dorn LE, Kim P, Sargent MA, Gunaje J, Otsu K, Davis JM, Circulation 2017, 116, 26238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Blaauboer ME, Smit TH, Hanemaaijer R, Stoop R, Everts V, Biochem. Biophys. Res. Commun 2011, 404, 23. [DOI] [PubMed] [Google Scholar]
- [66].Galie PA, Russell MW, Westfall MV, Stegemann JP, Exp. Cell Res 2012, 318, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Hinz B, Mastrangelo D, Iselin CE, Chaponnier C, Gabbiani G, Am. J. Pathol 2001, 159, 1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Hinz B, Nat. Med 2013, 19, 1567. [DOI] [PubMed] [Google Scholar]
- [69].Atance J, Yost MJ, Carver W, J. Cell. Physiol 2004, 200, 377. [DOI] [PubMed] [Google Scholar]
- [70].Schroer AK, Merryman WD, J. Cell Sci 2015, 128, 1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Sawada Y, Nakamura K, Doi K, Takeda K, Tobiume K, Saitoh M, Morita K, Komuro I, De Vos K, Sheetz M, Ichijo H, J. Cell Sci 2001, 114, 1221. [DOI] [PubMed] [Google Scholar]
- [72].Brault JJ, Pizzimenti NM, Dentel JN, Wiseman RW, J. Cell. Biochem 2013, 114, 1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Foster WH, Tidball JG, Wang Y, Muscle Nerve 2012, 45, 266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].MacKenna DA, Dolfi F, Vuori K, Ruoslahti E, J. Clin. Invest 1998, 101, 301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Hsu HJ, Lee CF, Locke A, Vanderzyl SQ, Kaunas R, PLoS One 2010, 5, e12470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Wang J, Seth A, McCulloch CA, Am. J. Physiol 2000, 279, H2776. [DOI] [PubMed] [Google Scholar]
- [77].Buscemi L, Ramonet D, Klingberg F, Formey A, Smith-Clerc J, Meister JJ, Hinz B, Curr. Biol 2011, 21, 2046. [DOI] [PubMed] [Google Scholar]
- [78].Wipff PJ, Rifkin DB, Meister JJ, Hinz B, J. Cell Biol 2007, 179, 1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Jenkins RG, Su X, Su G, Scotton CJ, Camerer E, Laurent GJ, Davis GE, Chambers RC, Matthay MA, Sheppard D, J. Clin. Invest 2006, 116, 1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Tomasek JJ, Martin MD, Vaughan MB, Cowan RL, Kropp BP, Mol. Biol. Cell 2000, 11, 88a. [Google Scholar]
- [81].Li Q, Xu Y, Li X, Guo Y, Liu G, Toxicol. Lett 2012, 211, 91. [DOI] [PubMed] [Google Scholar]
- [82].Zhang YM, Bo J, Taffet GE, Chang J, Shi J, Reddy AK, Michael LH, Schneider MD, Entman ML, Schwartz RJ, Wei L, FASEB J 2006, 20, 916. [DOI] [PubMed] [Google Scholar]
- [83].Meyer-ter-Vehn T, Han H, Grehn F, Schlunck G, Invest. Ophthalmol. Visual Sci 2011, 52, 9149. [DOI] [PubMed] [Google Scholar]
- [84].Huang X, Yang N, Fiore VF, Barker TH, Sun Y, Morris SW, Ding Q, Thannickal VJ, Zhou Y, Am. J. Respir. Cell Mol. Biol 2012, 47, 340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Godbout C, Follonier Castella L, Smith EA, Talele N, Chow ML, Garonna A, Hinz B, PLoS One 2013, 8, e64560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Pelham RJ Jr., Wang Y, Proc. Natl. Acad. Sci. USA 1997, 94, 13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Ko KS, Arora PD, McCulloch CA, J. Biol. Chem 2001, 276, 35967. [DOI] [PubMed] [Google Scholar]
- [88].Goffin JM, Pittet P, Csucs G, Lussi JW, Meister JJ, Hinz B, J. Cell Biol 2006, 172, 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Bell E, Ivarsson B, Merrill C, Proc. Natl. Acad. Sci. USA 1979, 76, 1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Tomasek JJ, Haaksma CJ, Eddy RJ, Vaughan MB, Anat. Rec 1992, 232, 359. [DOI] [PubMed] [Google Scholar]
- [91].Yang C, Tibbitt MW, Basta L, Anseth KS, Nat. Mater 2014, 13, 645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Mosqueira D, Pagliari S, Uto K, Ebara M, Romanazzo S, Escobedo-Lucea C, Nakanishi J, Taniguschi A, Franzese O, Di Nardo P, Goumans MJ, Traversa E, Pinto-do OP, Aoyagi T, Forte G, ACS Nano 2014, 8, 2033. [DOI] [PubMed] [Google Scholar]
- [93].Lin Z, von Gise A, Zhou P, Gu F, Ma Q, Jiang J, Yau AL, Buck JN, Gouin KA, van Gorp PR, Zhou B, Chen J, Seidman JG, Wang DZ, Pu WT, Circ. Res 2014, 115, 354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Xin M, Kim Y, Sutherland LB, Murakami M, Qi X, McAnally J, Porrello ER, Mahmoud AI, Tan W, Shelton JM, Richardson JA, Sadek HA, Bassel-Duby R, Olson EN, Proc. Natl. Acad. Sci. USA 2013, 110, 13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Kshitiz J. Park, Kim P, Helen W, Engler AJ, Levchenko A, Kim DH, Integr. Biol 2012, 4, 1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Chaterji S, Kim P, Choe SH, Tsui JH, Lam CH, Ho DS, Baker AB, Kim DH, Tissue Eng., Part A 2014, 20, 2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Beussman KM, Rodriguez ML, Leonard A, Taparia N, Thompson CR, Sniadecki NJ, Methods 2016, 94, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Kim P, Yuan A, Nam KH, Jiao A, Kim DH, Biofabrication 2014, 6, 024112. [DOI] [PubMed] [Google Scholar]
- [99].Kim DH, Han K, Gupta K, Kwon KW, Suh KY, Levchenko A, Biomaterials 2009, 30, 5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Huang C, Fu X, Liu J, Qi Y, Li S, Wang H, Biomaterials 2012, 33, 1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Rouillard AD, Holmes JW, Biophys. J 2014, 106, 932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Booth AJ, Hadley R, Cornett AM, Dreffs AA, Matthes SA, Tsui JL, Weiss K, Horowitz JC, Fiore VF, Barker TH, Moore BB, Martinez FJ, Niklason LE, White ES, Am. J. Respir. Crit. Care Med 2012, 186, 866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Ali SR, Ranjbarvaziri S, Talkhabi M, Zhao P, Subat A, Hojjat A, Kamran P, Muller AM, Volz KS, Tang Z, Red-Horse K, Ardehali R, Circ. Res 2014, 115, 625. [DOI] [PubMed] [Google Scholar]
- [104].Kanisicak O, Khalil H, Ivey MJ, Karch J, Maliken BD, Correll RN, Brody MJ, Lin SCJ, Aronow BJ, Tallquist MD, Molkentin JD, Nat. Commun 2016, 7, 12260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Pagliari S, Mosqueira D, Escobedo-lucea C, Goumans MJ, Pinto-do-o P, Aoyagi T, Forte G, Tissue Eng J. Regener. Med 2014, 8, 178. [Google Scholar]