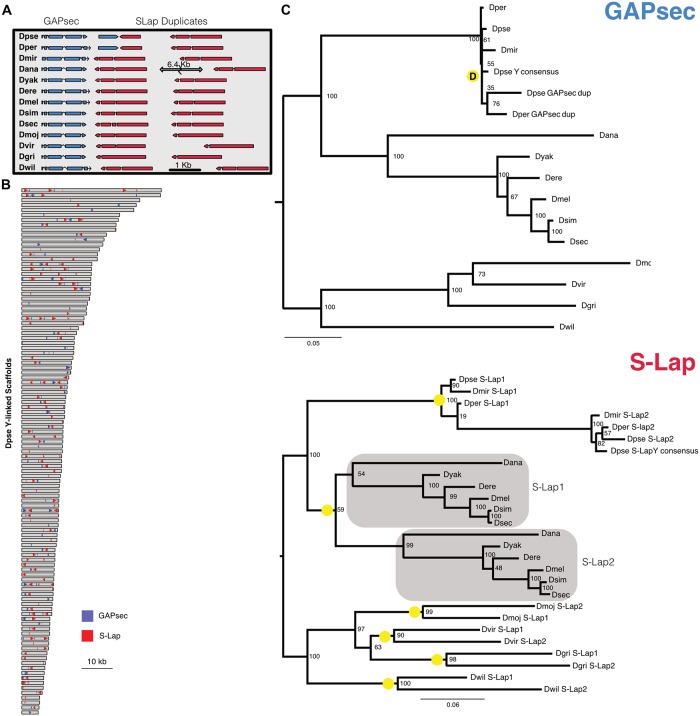
Fig 2. Molecular evolution of the S-Lap1 and GAPsec genes in Drosophila.
A. Organization of S-Lap1 and GAPsec genes across the Drosophila genus. S-Lap1 (shown in red) is duplicated in all Drosophila species investigated, and GAPsec (shown in blue) shows a partial duplication in D. pseudoobscura and its sister species D. persimilis. B. Amplification of S-Lap1 and GAPsec on Y-linked scaffolds of D. pseudoobscura. Each rectangle represents a Y-linked genomic scaffold whose size is proportional to the scaffold length. The location and orientation of S-Lap1 (red) and GAPsec (blue) duplicates on each scaffold is represented by the colored arrowheads. C. Gene trees of S-Lap1 and GAPsec copies. A RAxML maximum likelihood phylogeny was inferred from alignments of the GAPsec and S-Lap transcripts for 12 Drosophila species plus D. miranda. The conserved genomic location of S-Lap1 and S-Lap2 across Drosophila suggests that they duplicated in an ancestor of Drosophila, and that gene conversion (indicated by the yellow circles) homogenized these tandem duplicates at different branches along the phylogeny. An inferred duplication event of GAPsec is shown by the yellow circle with a D. Alignments used to generate the trees shown in Fig 1C are provided as S1 and S2 Data.