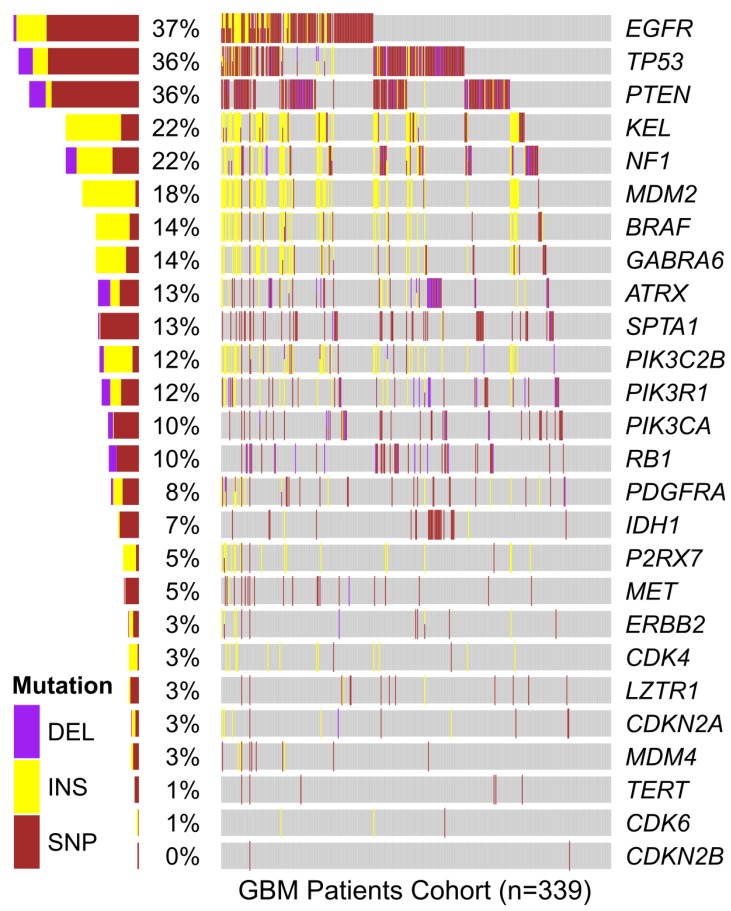
Figure 8. In silico analysis of genetic mutations found in GBM samples from the TCGA cohort.
The presence of mutations in the genes listed vertically is specified by color for each of the 339 individuals in the cohort (represented horizontally along the gray bar). Total percentage of deletions (DEL), insertions (INS) and single nucleotide polymorphisms (SNP) verified in the P2RX7 gene and other genes mutated in GBM is specified on the left side. In P2RX7, two types of missense mutation (g.chr12:121593912C>G and g.chr12:121603192C>T), one type of splice site (g.chr12:121613190G>T), one type of silent (g.chr12:121622170G>A) and three types of frameshift insertions (rs61745024; g.chr12:121613270_121613271insT and g.chr12:121622164_121622165insC) were identified. Data were analyzed using a free software environment for statistical computing and graphics (R Project).