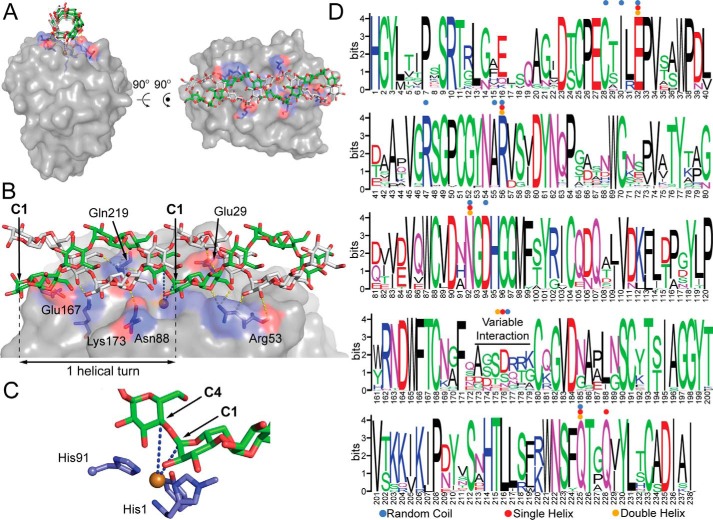
Figure 7.
Docking model of an amylose double helix bound to AoAA13. A, illustration of the amylose double helix bound to the active-site surface. B, residues identified in the model to make hydrogen bonds (yellow dashed lines) with amylose. This model was generated from a docking experiment using PDB code 4OPB. C, close-up of the active site. The blue dashed lines indicate the distance from the copper center (brown) to the C1 (5.1 Å) and C4 (5.8 Å) positions of the glycosidic linkage positioned over the active site. D, sequence logo for the catalytic domain of the putative AA13 PMO family, showing conserved contact sites for modeled substrates: random (blue dots), single helix (red dots), or double helix (orange dots) amylose. All binding sites predicted in the docking model involve highly conserved residues in the AA13 family, with the exception of the Glu167/Pro172/Lys173 contact sites, which are found in a highly variable region of the sequence. Gaps in the logo caused by nonconserved insertions were manually removed for clarity. Numbers do not exactly match the sequence numbering of NcAA13, as many other sequences in the family have relatively conserved insertions.