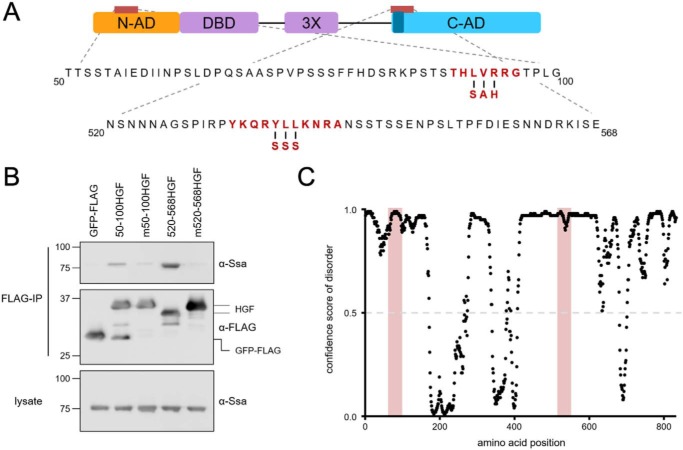
Figure 3.
Ssa1-binding sites are required for Ssa1 to interact with the N-AD and C-AD. A, mutations designed to disrupt the LIMBO algorithm-predicted potential Hsp70-binding site in the N-AD and the known CE2 site (dark blue) in the C-AD are highlighted in red. B, Ssa1 requires intact Hsp70-binding sites to associate with the indicated N-AD or C-AD constructs described previously in the legend to Fig. 2, as demonstrated by failure to bind fusions bearing the indicated mutations (designated m50–100HGF and m520–568HGF). Immunoblots are representative of three independent experiments. C, both the N-AD and C-AD are predicted to be intrinsically disordered, with confidence scores for disorder above 0.5, including the Hsp70-binding sites highlighted in light red. Disorder was predicted by the DISOPRED3 algorithm (35).