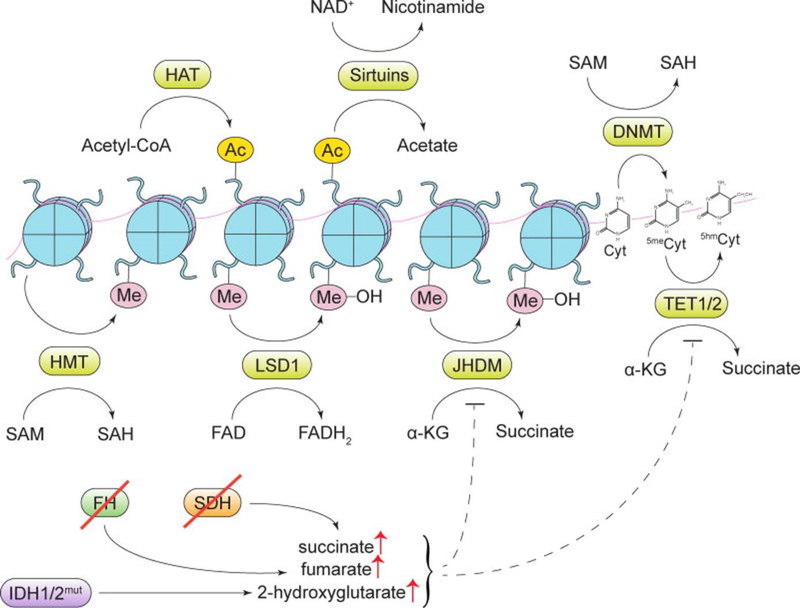
Figure 3. Cellular metabolites contribute to gene regulation and their fluctuating levels affect the epigenetic process.
Addition or removal of epigenetic marks by several key epigenetic enzymes is dependent on metabolites that act as substrates or cofactors of these enzymes. [HAT, histone acetyltransferase enzymes; Ac, an acetyl mark; SAM, S-adenosylmethionine; SAH, S-adenosylhomocysteine; DNMT, DNA methyltransferase enzymes; HMT, histone methyltransferase enzymes; Me, a methyl mark; LSD1, lysine-specific histone demethylase 1; JHDM, Jumonji domain-containing histone demethylase enzymes; Cyt, cytosine; 5meCyt, 5-methylcytosine; 5hmCyt, 5-hydroxymethylcytosine; TET1/2, ten-eleven translocation methylcytosine dioxygenase 1/2; α-KG, α-ketoglutarate; SDH, succinate dehydrogenase; FH, fumarate hydratase; IDH1/2, isocitrate dehydrogenase 1/2]. Image is adapted from[92]