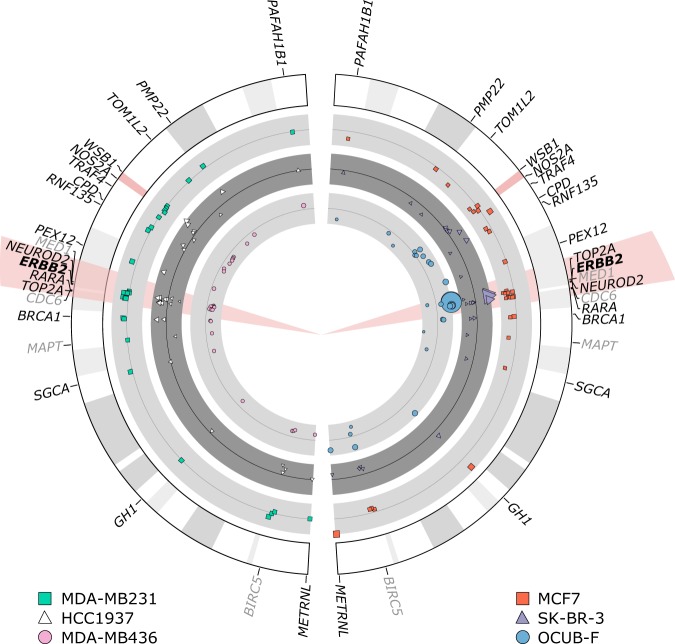
Figure 2.
MLPA results of chromosome 17 of all cell lines used. Cell lines MDA-MB231, HCC1937, MCF7, SK-BR-3, OCUB-F and MDA-MB436 are shown. Probemix P004-B1 and P078-B1 (greytone) were used on all cell lines. The HER2 gene is highlighted in red. The size of the used pictograms corresponds to the reach of the MLPA results (bigger size correlates to higher reach). Results between the probemixes were concordant. The P078-B1 breast tumour probemix also contained probes for genes located on other chromosomes relevant for breast carcinoma and reference genes. In these genes no polysomy or loss or amplification could be detected (not shown in the figure). Negative controls used, were nicely oriented around 1,00 (not shown). The MLPA results with probemix P004-B1 and P078-B1 (greytone) of cell lines MDA-MB231 and HCC1937 show no polysomy or loss or amplification of HER2. MCF7 shows partial loss of the HER2 gene, corresponding to FISH results (Table 1). SK-BR-3 shows amplification of the HER2 gene corresponding to IHC results and FISH results (Fig. 1). OCUB-F shows amplification of the HER2 gene corresponding to the IHC and FISH results (Table 1). MDA-MB436 shows partial loss of all tested genes on chromosome 17, concordant with monosomy, as was seen with FISH (Fig. 1).