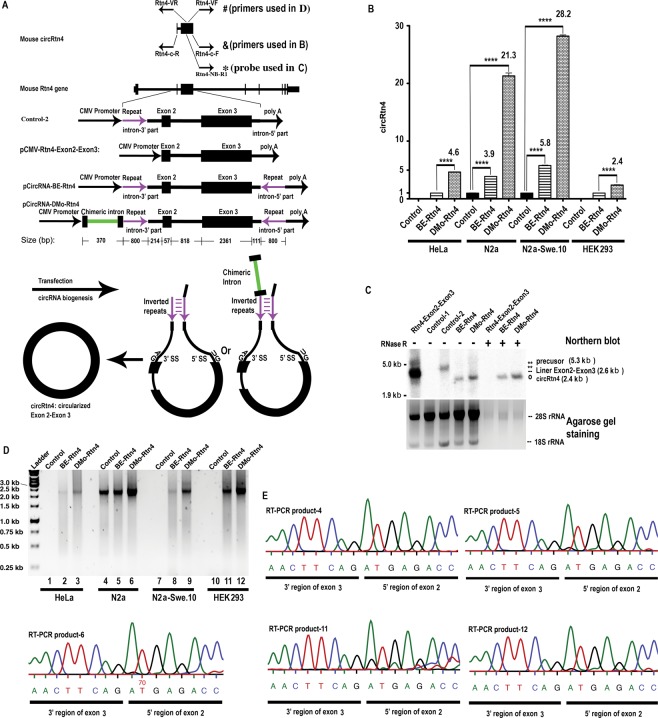
Figure 1.
Mouse circRtn4 structure and generation in mammalian cells. (A) Schematic representation of the circRtn4 localization within the mouse Rtn4 gene environment and the circRNA cassette for pCircRNA-BE and pCircRNA-DMo vectors; mouse circRtn4 consists of Rtn4 gene exons 2 and 3. The 800 nt inverted repeats (purple colour) within the flanking introns were inserted to promote backsplicing through the formation of inter-intronic base-pairing interactions; flanking introns lack 5′ and 3′ splice sites, which lead to abolished canonical splicing of exon 2 and exon 3; the chimeric intron is displayed in green; & and # indicate the circRtn4 RT-PCR oligonucleotide positions; &: Rtn4-c-R and Rtn4-c-F were used for qRT-PCR to determine circRtn4 levels (results shown in B); #, Rtn4-VR, Rtn4-VF were used for the analysis of circRtn4 backsplicing fidelity (results shown in C); *indicates the position of the oligonucleotide probe for Northern blot hybridization (Rtn4-NB-R1) as displayed in D. (B) circRtn4 levels in transfected cells (HeLa, N2a, N2a-swe.10, HEK293 cell); pCMV-MIR empty vector as negative control; BE-Rtn4, pCircRNA-BE-Rtn4; DMo-Rtn4, pCircRNA-DMo-Rtn4; all T-tests were performed in comparison to levels of the control sample, ****P ≤ 0.0001, n ≥ 4; β-actin mRNA was used as internal control. (C) Northern blot hybridization for detection of circRtn4 in transfected HEK293 cells. Control-1, pCMV-MIR empty vector; Rtn4-Exon2-Exon3, pCMV-Rtn4-Exon2-Exon3; Control-2, the construct devoid of the downstream portion of the inverted repeat in the 3′ flanking intronic region; BE-Rtn4, pCircRNA-BE-Rtn4; DMo-Rtn4, pCircRNA-DMo-Rtn4; −, no RNase R treatment; +, with RNase R treatment; agarose gel ethidium bromide staining of 28S and 18S rRNAs served as loading control; the weak staining of 18S rRNA is due to co-migration and signal quenching with xylene cyanol loading dye. (D) Agarose gel electrophoresis of RT-PCR products of circRtn4 to analyse backsplicing fidelity (PCR primers: Rtn4-VR, Rtn4-VF); pCMV-MIR empty vector as negative control; BE-Rtn4, pCircRNA-BE-Rtn4; DMo-Rtn4, pCircRNA-DMo-Rtn4. The entire un-spliced precursor transcript is ~5.3 kb; the spliced circular isoform of exons 2 and exon 3 devoid of the intron corresponds to 2.4 kb. The intron flanked by exons 2 and 3 is 818 nt long. The expected amplicon size is 2.4 kb. Products migrated between 2.0 and 2.5 kb, indicating that the internal intron was spliced during circRtn4 biogenesis. Lane 1–3, HeLa cells; lane 4–6, N2a cells; lane 7–9, N2a-swe.10 cells; lane 10–12, HEK293 cells. PCR products were sequenced and aligned (data not shown). (E) Sequencing of the junction site for circRtn4 backsplicing as revealed by assays in N2a and HEK293 cell lines. The RT-PCR products as displayed in C were sequenced and the junction regions were shown; RT-PCR product-4, 5, 6 were from N2a cells; RT-PCR product-11, 12 were from HEK293 cells.