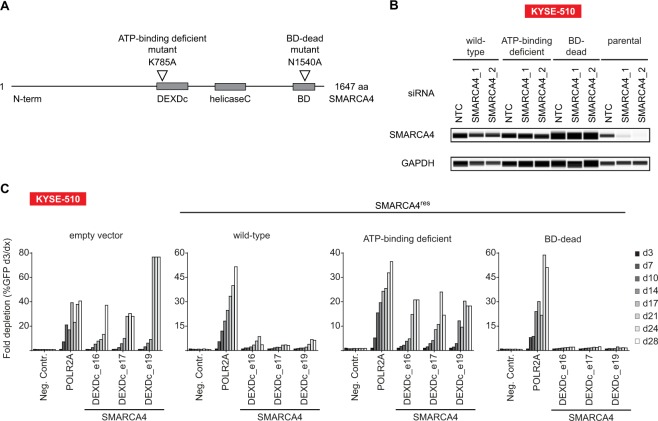
Figure 3.
SMARCA4 dependency in SMARCA2-deficient ESCC cell lines is linked to its ATPase function. (A) Schematic representation of SMARCA4 domain structure. Location of ATP-binding- and bromodomain (BD)-inactivating mutations in siRNA/sgRNA-resistant SMARCA4 (SMARCA4res) is indicated. DEXDc, DEAD-like helicases superfamily; BD, bromodomain. (B) SMARCA2-deficient KYSE-510 cells were stably transduced with wild-type or mutant forms of SMARCA4res. Transgene expression was monitored by capillary Western immunoassay in presence of siRNA-mediated knockdown of endogenous SMARCA4. Lysates were prepared 72 h post siRNA transfection. GAPDH expression was used to monitor equal loading. (C) Time-resolved CRISPR-Cas9 depletion studies in SMARCA4res expressing KYSE-510 cells. Fold depletion of sgRNA- and GFP-co-expressing cells relative to day 3 (d3) post-transduction was assessed by flow cytometry analysis over 28 days (n = 1 experimental replicate).