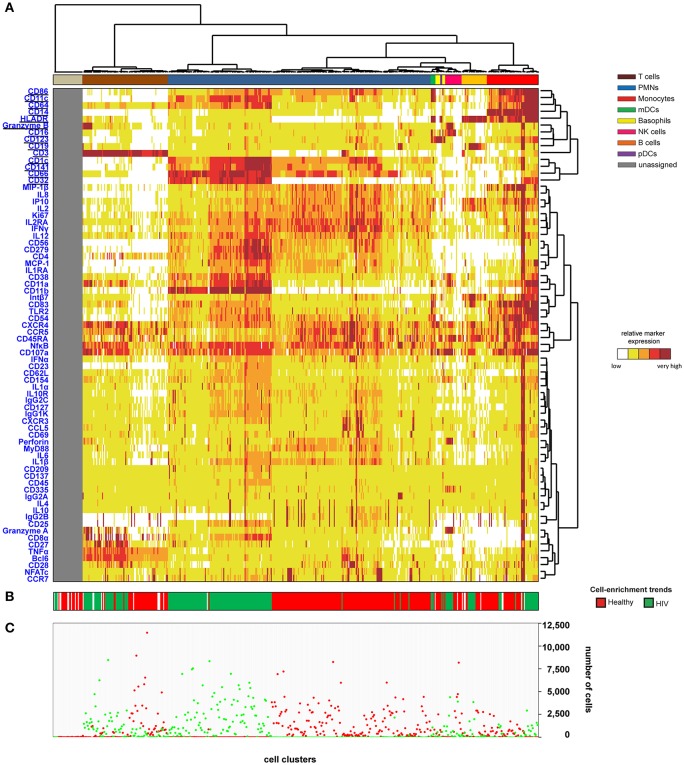
Figure 1.
Heatmap representing immune cells from healthy and HIV-infected individuals. Whole blood was collected from three non-viremic HIV-infected patients and three healthy subjects. Following red blood cell lysis, leukocytes of each sample were stained using three mass cytometry panels consisting of 35, 32, and 33 markers. Fourteen markers were shared between the three panels. CytoBackBone was used to generate a merged cytometric profile at the cellular level for each of the six individuals. SPADE analysis was performed to identify 500 cell clusters. The SPADE clustering was generated based on the expression levels of the whole set of 72 cell markers. (A) A categorical heatmap representing the relative marker expression for each cluster was generated. Marker expression ranges were calculated based on the 5 and 95th percentile of marker expression. Then, each range was divided into five uniform categories. The categorization of marker expression was computed based on the means of the individual SPADE expression medians of each marker. The categories represent negative, low, medium, high, and bright relative marker expression using a color scale from white to dark red. Hierarchical clustering was performed to visualize clusters with similar expression patterns. Two additional hierarchical clusterings were performed to visualize markers with similar expression patterns: one for common markers and one for non-common markers. Underlined markers correspond to the backbone markers (i.e., markers present in all cytometric panels). Cell clusters were manually annotated on the heatmap. Colors correspond to cell populations. (B) The cell-enrichment trend, toward HIV or healthy profiles, is indicated for each cluster. (C) The sum of the cells for all individuals and each condition is indicated for each cluster.