Fig. 1.
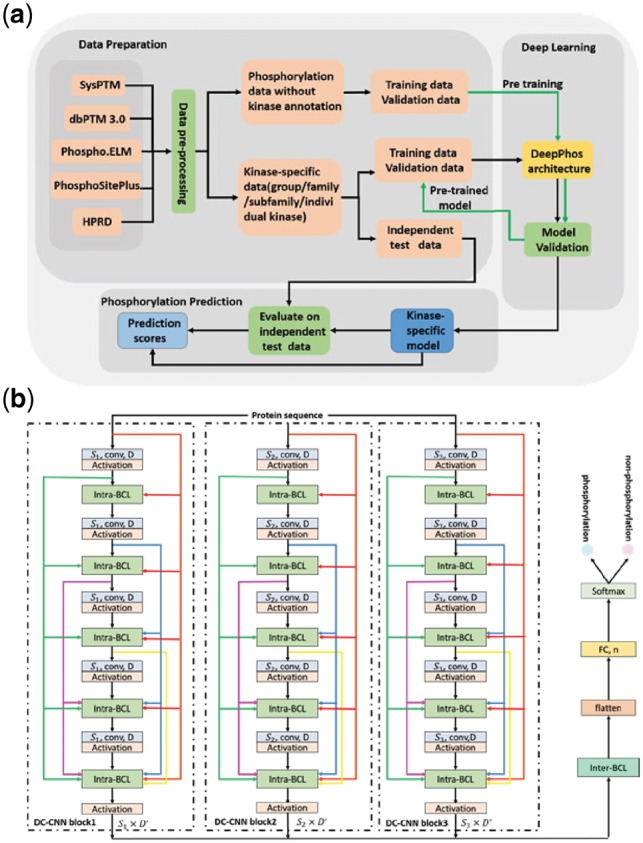
DeepPhos overall framework. (a) The working flow of kinase-specific phosphorylation site prediction in DeepPhoos. (b) The network layout of the DeepPhos approach. The raw sequences are transformed into a set of sequence features by one-hot codings. Intra-BCLs between two convolutional layers in each DC-CNN block are designed for connecting previous and current feature maps as shown by the different colorful arrows, in which feature maps are concatenated in the feature dimension. in the convolutional layers refers to the width of the filters used in convolutional operation, D represents the number of filters used in convolutional layers, and refers to the size of output feature maps. The representations generated by multi-blocks are further integrated by inter-BCL and transformed to FC layer (n represents the number of neurons), and finally generate the phosphorylation prediction by softmax function (Color version of this figure is available at Bioinformatics online.)
