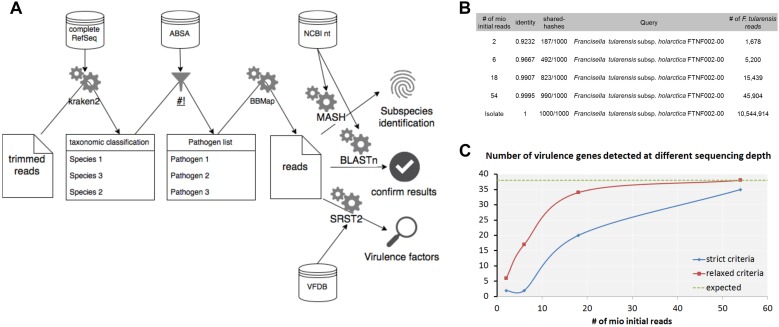
FIGURE 9.
Workflow for the detection and characterization of pathogenic microorganisms using shotgun metagenomics sequencing. Taxonomic profiles are generated with kraken2 using the complete RefSeq database. The resulting taxonomic abundance table is filtered for pathogenic species using ABSA database. For detected pathogens classified reads are extracted and verified with BLASTn using the nt database from NCBI. Subspecies is resolved by determination of closest available reference using Mash. Virulence factors are detected with SRST2 in combination with the VFDB. (A) Analysis of a hare liver sample infected with F. tularensis subsp. holarctica using the workflow shows determination of the F. tularensis subspecies (B) and the detection of virulence factors (C) at different read depths from metagenomic sequencing data and WGS data of the isolate extracted from the same hare liver sample.