Figure 3. CHROME depletion in hepatocytes reduces cholesterol efflux and formation of nascent HDL particles.
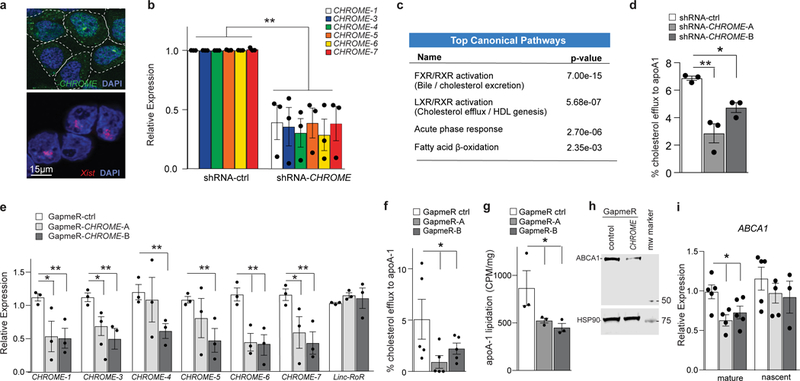
a, Fluorescence in situ hybridization for CHROME (green) and Xist (red) RNA in HEK293T cells stained with DAPI to visualize nuclear DNA (blue). b, qPCR analysis of CHROME variants in HepG2 cells stably expressing an shRNA targeting all CHROME variants (shRNA-CHROME) or control shRNA (shRNA-ctrl). c, Ingenuity pathway analysis of RNA-Seq data from shRNA-CHROME and shRNA-ctrl expressing HepG2 cells showing the top canonical pathways altered upon CHROME knockdown. d, Measurement of cholesterol efflux to exogenous apoA-1 in HepG2 cells expressing shRNA-CHROME or shRNA-ctrl. e, qPCR quantification of CHROME variants in primary human hepatocytes transfected with control (GapmeR-ctrl) or CHROME-targeting GapmeRs (GapmeR-CHROME-A or GapmeR-CHROME-B). f, Measurement of cholesterol efflux to exogenous apoA-1 in primary human hepatocytes transfected with control or CHROME-targeting GapmeRs. g, Measurement of phospholipid efflux to apoA1 secreted from primary human hepatocytes labelled with C14-choline chloride and transfected with control or CHROME-targeting GapmeRs. h, Western blot analysis of ABCA1 and HSP90 (control) in hepatocytes transfected with control or CHROME-targeting GapmeRs. Molecular weight marker is shown at right. i, qPCR analysis of mature and nascent ABCA1 mRNA transcripts in primary human hepatocytes transfected with control or CHROME-targeting GapmeRs. b, d-i, Data are the mean ± SEM of 3–5 independent experiments. P-values were calculated using a two-tailed student’s t-test. *P≤0.05, **P≤0.01.
