Figure 5. Gain or loss of CHROME in hepatocytes alters levels of its interacting miRNAs and their common target mRNAs involved in cholesterol metabolism.
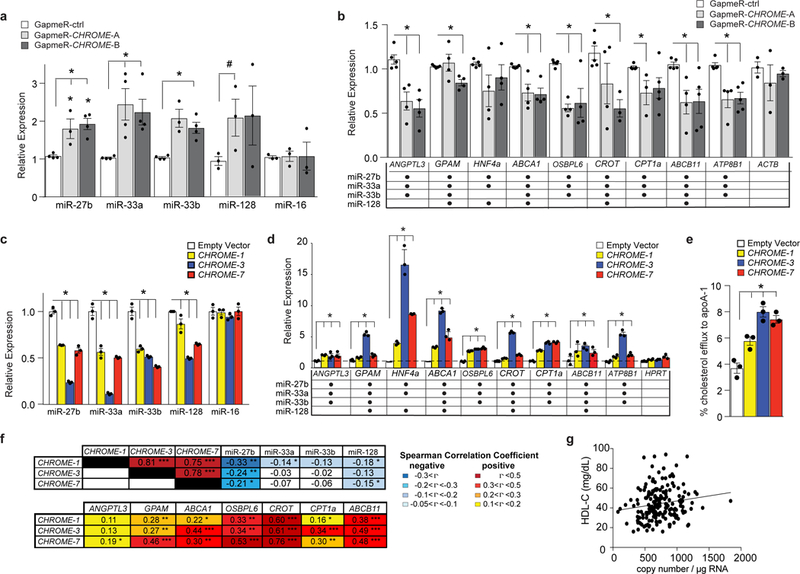
a-d, qPCR analysis of levels of (a, c) miRNAs and (b, d) their common target mRNAs in (a-b) primary human hepatocytes transfected with control (GapmeR-ctrl) or CHROME-targeting GapmeRs (GapmeR-CHROME-A or GapmeR-CHROME-B) or (c-d) HepG2 cells stably expressing CHROME-1, CHROME-3, CHROME-7 or empty vector. Table in (b) and (d) indicates validated targets of miR-27b, miR-33a, miR-33b and miR-128. e, Measurement of cholesterol efflux to exogenous apoA-1 in THP-1 macrophages stably expressing empty vector, CHROME-1, CHROME-3 or CHROME-7. f, Spearman rank correlation coefficients (rho) of CHROME RNA copy number with its interacting miRNAs and their target mRNAs in human liver (n=200). g, Scatter plot showing relationship between hepatic CHROME-7 copy number and plasma levels of HDL cholesterol (n=200). Spearman correlation coefficients between CHROME-7 copy number and plasma levels of HDL cholesterol (n=200, rho=0.1722, P=0.0019) were calculated using the SAS statistic software. Data in (a-e) are the mean ± SEM of 3 independent experiments and P-values were calculated using a two-tailed student’s t-test. #P≤0.1, *P≤0.05, **P≤0.001, ***P≤0.000001.
