Figure 6. CHROME regulates cholesterol efflux in macrophages via its interaction with miRNAs.
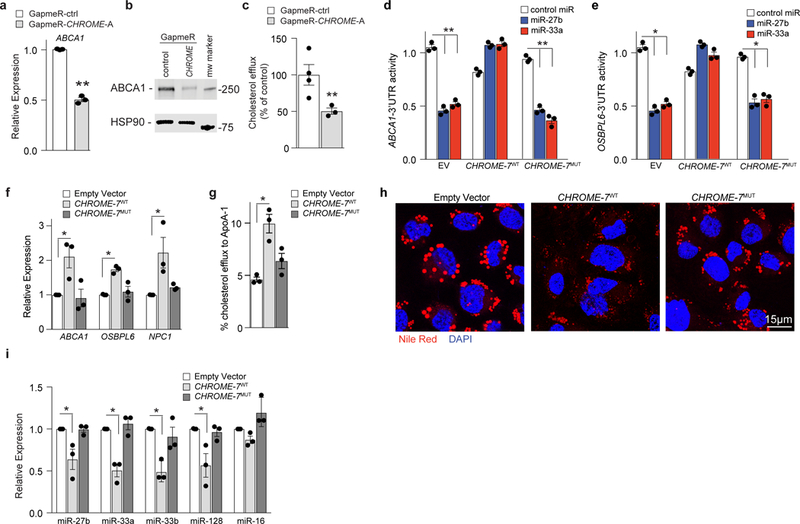
a) qPCR analysis of ABCA1 mRNA in THP-1 macrophages transfected with control (ctrl) or CHROME-targeting GapmeRs. Data are the mean ± SEM of 3 independent experiments. (b) Western blot of ABCA1 and HSP90 (internal control) protein in THP-1 macrophages transfected with control (ctrl) or CHROME-targeting GapmeRs. Molecular weight marker is shown at right. Data are representative of 3 independent experiments. c, Measurement of cholesterol efflux to exogenous apoA-1 in THP-1 macrophages transfected control (ctrl) or CHROME-targeting GapmeRs. Data are the mean ± SEM of 4 independent experiments. d-e, Activity of (d) ABCA1-3’UTR and (e) OSBPL6-3’UTR luciferase reporter genes in HEK293T cells expressing empty vector (EV), wild type CHROME-7 (CHROME-7WT), or CHROME-7 with its miR-33, miR-27b and miR-128 binding sites mutated (CHROME-7MUT) following treatment with control miR, miR-27b or miR-33 mimics. Data are the mean ± SEM of 3 biological replicates from a single experiment. Data are representative of 3 independent experiments. f, qPCR analysis of miR-27b, miR-33 and miR-128 target genes regulating cholesterol efflux in THP-1 macrophages stably expressing empty vector, CHROME-7WT or CHROME-7MUT. Data are the mean ± SEM of 3 experiments. g, Measurement of cholesterol efflux to apoA-1 in THP-1 macrophages stably expressing empty vector, CHROME-7WT or CHROME-7MUT. Data are the mean ± SEM of 3 experiments. h, Representative images of Nile Red stained lipid droplet accumulation in acetylated LDL treated THP-1 macrophages stably expressing empty vector, CHROME-7WT or CHROME-7MUT. Cells were stained with DAPI (blue) to visualize nuclear DNA. i, qPCR analysis of miRNA levels in THP-1 cells stably expressing empty vector, CHROME-7WT or CHROME-7MUT. miR-16 is included as a negative control. Data are the mean ± SEM of 3 experiments. P-values were calculated using a two-tailed student’s t-test. *P≤0.05, **P≤0.01, ***P≤0.001.
