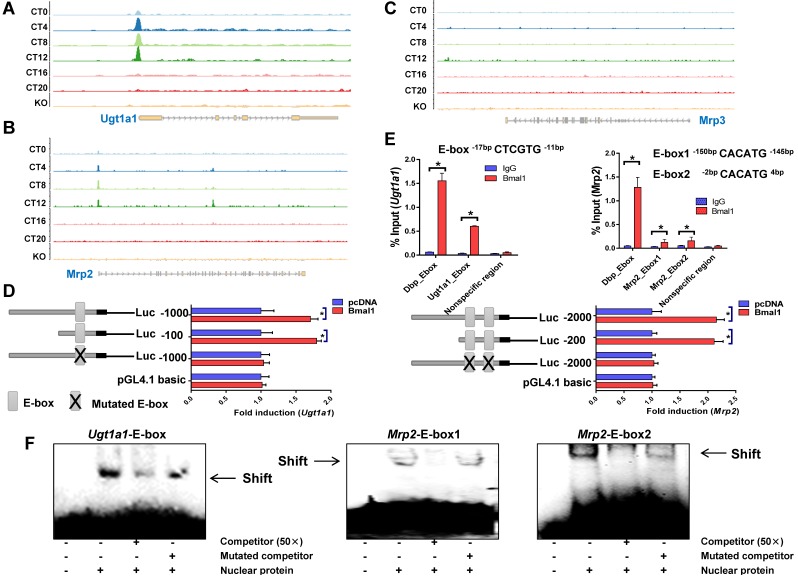
Figure 4.
Bmal1 regulates Ugt1a1 and Mrp2 transcription. ChIP-sequencing for circadian Bmal1 binding to Ugt1a1 (A), Mrp2 (B) and Mrp3 (C). Peaks indicate regions of DNA bound by Bmal1. ChIP-sequencing traces were generated from GSE39977. (D) Luciferase reporter assays with Hepa1-6 cells, showing the effects of Bmal1 on the activities of different versions of Ugt1a1 promoters [i.e., Ugt1a1 (-1000-bp~+100-bp), Ugt1a1 (-100-bp~+100-bp) and Ugt1a1 mutant (-1000-bp~+100-bp)], Mrp2 promoters [i.e., Mrp2 (-2000-bp~+100-bp), Mrp2 (-200-bp~+100-bp) and Mrp2 mutant (-2000-bp~+100-bp)] and pGL4.1 basic vector. Data are mean ± SD (n=6). *P < 0.05 (t test). (E) ChIP assays showing interactions of Bmal1 with Ugt1a1 and Mrp2 promoters in the livers of wild-type mice at ZT2. Immunoprecipitated chromatin was measured by qPCR with primers specific for the regions of Dbp, Ugt1a1 and Mrp2. Data are mean ± SD (n=5). *P < 0.05 (t test). (F) EMSA assays indicating that Bmal1 binds to E-boxes in the region of Ugt1a1 promoter (-17-bp~-12-bp) and in the region of Mrp2 promoter (-150-bp~-145-bp and -2-bp ~+4-bp).