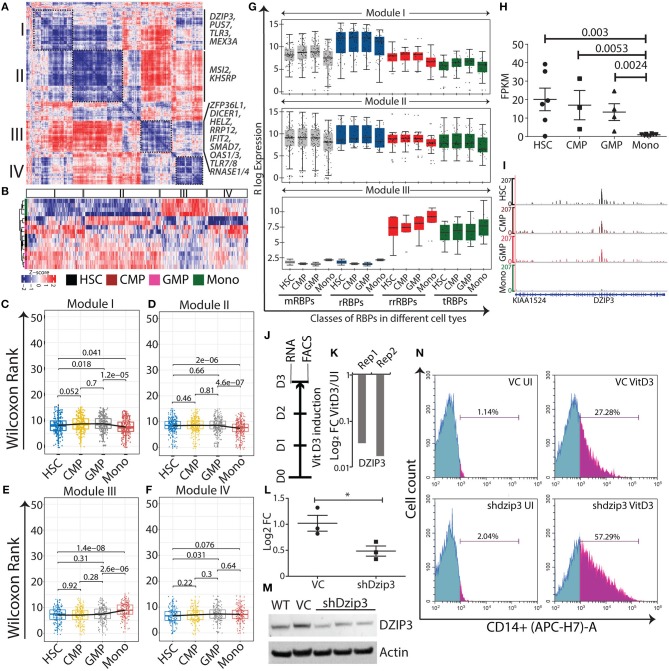
Figure 3.
Normal myeloid cell-specific modules obtained from gene-gene correlation studies. Heat map showing Spearman correlation plot based on RBP gene expression values, and the black boxes show four modules that were considered for further studies (A). Heat map representation of unsupervised clustering of RBP gene expression in different modules (B). Box-whisker plots represent overall gene expression patterns of RBPs in HSC, CMP, GMP, and monocytes across four modules. Wilcoxon rank test shows p-values for each comparisons (C–F). Box-whisker plots represent the gene expression pattern of RBP classes within modules I, II, and III; p-values for comparing the difference in each RBP class for different cell types is listed in Supplementary Table 5 (G). Expression profile of DZIP3 was plotted from the bloodspot database using BLUEPRINT RNA-seq data (H). Genome browser plots of DZIP3 from RNA-seq data analyzed in this paper (I). Experimental workflow to identify the role of DZIP3 in U937 promyelocyte differentiation (J). U937 cells were treated with 30 nM vitamin D3 for 72 h and DZIP3 transcript levels were assessed using qRT-PCR and plotted relative to actin levels in two biological replicates (K). U937 cells were transduced with lentivirus containing shRNA against DZIP3, and 48 h post-puromycin selection, knockdown was confirmed both at the transcript level using qRT-PCR (relative to actin) (L) and at protein level using immunoblotting; actin was used as the loading control (M) from three independent experiments; error bars are mean ± s.e.m. *P < 0.05 per two-tailed Student's t-test. Representative histograms showing expression of monocyte-specific marker CD14 of three independent experiments. CD14 expression was assessed using flow cytometry, after vitamin D3 induction for 72 h in, vector control (VC) U937 (upper panel), and shdzip3 transduced U937 (lower panel) (N).