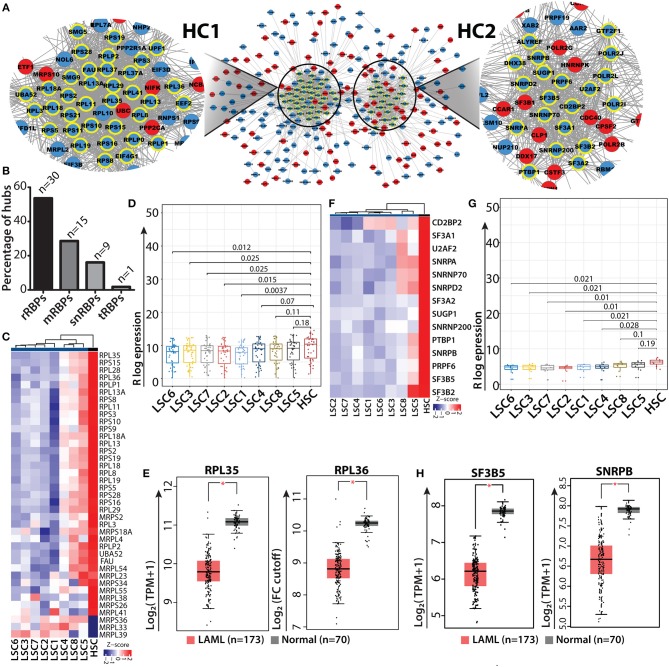
Figure 5.
RBP interaction network and ribosomal RBP hubs in LSC. Shortest path gene interaction network of RBP and non-RBPs from module II; RBP hub genes are highlighted with bold yellow borders, and the color of the nodes depict the mode of expression of the gene in LSC compared to in HSC. Red denotes upregulated and blue indicates downregulated in LSC compared to HSC. Magnified view of a network depicting cluster of ribosomal proteins (hub-cluster 1 as HC1) and splicing related RBPs (hub-cluster 2 as HC2) (A). Bar plot showing the percentage of hubs for different classes of RBPs; “n” represents number of RBPs (B) Heat map showing ribosomal RBP gene expression (n = 30) across individual LSCs compared to in HSCs (averaged expression values) (C). Box-whisker plot (Wilcoxon rank test p-values) depicting overall downregulation of ribosomal proteins in LSCs (D). Box-whisker plots showing significant downregulation of ribosomal hub RBPs RPL35 and RPL36 in AML from TCGA, GTEx data cohort visualized in GEPIA2 using default parameters; (one-way ANOVA, p < 0.01) (E). Heat map and box-whisker plot (Wilcoxon rank test p-values) showing downregulation of splicing related RBPs (n = 14) in LSCs compared to HSCs (F,G). Box-whisker plots showing significant downregulation of splicing related hub RBPs SF3B5 and SNRPB in AML from TCGA, GTEx data cohort visualized in GEPIA2 using default parameters; (one-way ANOVA, p < 0.01) (H). *p < 0.01.