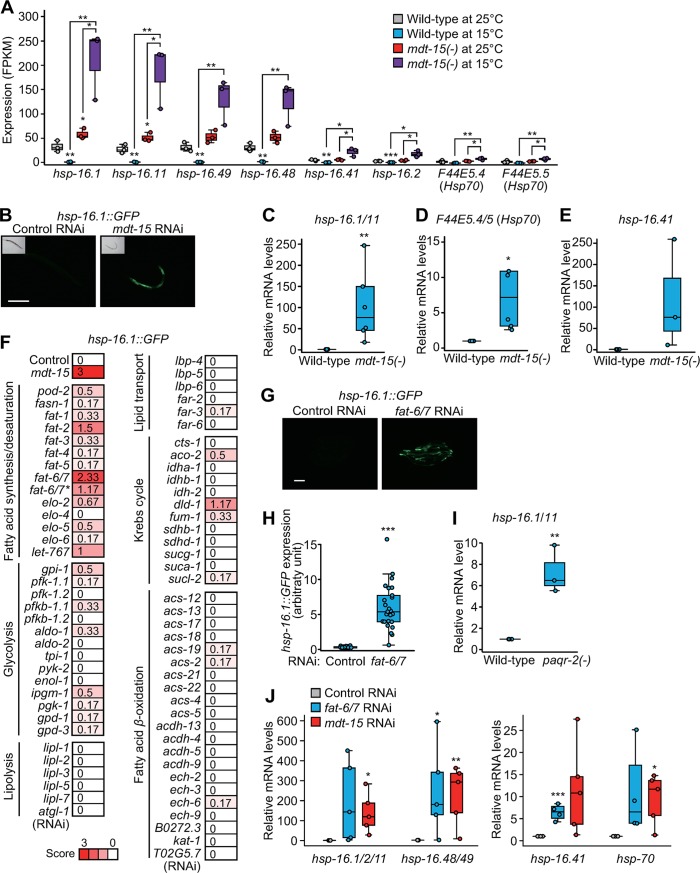
Fig 5. Decreased UFA/SFA ratio induces cytosolic chaperones.
(A) mRNA levels of selected cytosolic chaperones, hsp-16.1, hsp-16.11, hsp-16.49, hsp-16.48, hsp-16.41, hsp-16.2, F44E5.4, and F44E5.5 in wild-type and mdt-15(-) worms at 25 °C and 15 °C were measured by using RNA-seq (n = 3, two-tailed Student t tests, *p < 0.05, **p < 0.01). Note that the mRNA levels of hsp-16.1 and hsp-16.11, those of hsp-16.48 and hsp-16.49, and those of F44E5.4 and F44R5.5 may not be completely distinguishable because the sequences of these pairs of transcripts are very similar. The induction of some of these chaperone genes by inhibition of mdt-15 was also observed at 20 °C in a previous report [33]. See S1 Data and GEO database (accession number GSE128815) for RNA-seq data. (B) Images of hsp-16.1p::hsp-16.1::GFP [hsp-16.1::GFP] transgenic worms treated with control RNAi or mdt-15 RNAi at 15 °C. Scale bar: 200 μm. (C) mRNA levels of hsp-16.1/11 were measured by using qRT-PCR in wild-type and mdt-15(tm2182) [mdt-15(-)] animals at 15 °C (n = 6). (D) The expression of F44E5.4/5 at 15 °C was determined by using qRT-PCR (n = 5). (E) mRNA levels of hsp-16.41 in wild-type and mdt-15(-) at 15 °C (n = 3). The expression changes were not statistically significant because of large variations. (F) A targeted RNAi screen against metabolic genes using hsp-16.1::GFP. Heat maps display average arbitrary scores between 0 to +3 determined by two independent researchers from three different experimental sets. RNAi targeting each of genes encoding various enzymes that regulate fatty acid synthesis/desaturation, glycolysis, lipolysis, lipid transport, Krebs cycle, and fatty acid β-oxidation were tested. fat-6/7 RNAi clones are predicted to target both fat-6 and fat-7, which are 87% identical [74]. Further experimental data using fat-6/7* were excluded based on our qRT-PCR analysis (S5D Fig). (G) Representative fluorescence images of hsp-16.1::GFP treated with control RNAi or fat-6/7 RNAi at 15 °C. Knocking down fat-6 and fat-7 increased the level of hsp-16.1::GFP. Scale bar: 200 μm. (H) Quantification of the hsp-16.1::GFP treated with control RNAi or fat-6/7 RNAi at 15 °C. Images of the individual worms were obtained separately for the quantification. (n ≥ 23 from three independent experiments). (I) paqr-2(tm3410) [paqr-2(-)] mutations induced the expression of hsp-16.1/11 at 15 °C measured by using qRT-PCR (n = 3). (J) Relative expression of cytosolic chaperone genes by RNAi knock-down of fat-6/7 and mdt-15 at 15 °C (n ≥ 4, two-tailed Student t tests, *p < 0.05, **p < 0.01, ***p < 0.001). The effect of RNAi targeting fat-6/7 or mdt-15 on the expression of fat-6 and fat-7 are shown in S5D Fig. See S7 Table for the standard deviations of box plot data. ACDH, acyl-CoA dehydrogenase; ACS, acyl-CoA synthetase; aldo, aldolase; atgl, adipose triglyceride lipase; cts, citrate synthase; dld, dihydrolipoamide dehydrogenase; ECH, enoyl-CoA hydratase; ELO, fatty acid elongase; enol, enolase; FAR, fatty-acid–and retinol-binding protein; FAS, fatty acid synthase; FPKM, fragments per kilobase of exon model per million reads mapped; fum, fumarase; GEO, Gene Expression Omnibus; GFP, green fluorescent protein; gpd, glyceraldehyde 3-phosphate dehydrogenase; GPI, glucose-6-phosphate isomerase; hsp, heat shock protein; IDH, isocitrate dehydrogenase; ipgm, cofactor-independent phosphoglycerate mutase; kat, 3-ketoacyl-coA thiolase; LBP, lipid-binding protein; let, lethal; lipl, lipase-like; MDT-15, Mediator 15; paqr, progestin and adipoQ receptor; PFK, phospho-fructo-kinase; PGK, phosphoglycerate kinase; pod, polarity and osmotic sensitivity defect; pyk, pyruvate kinase; qRT-PCR, quantitative reverse-transcription PCR; RNAi, RNA interference; RNA-seq, RNA sequencing; SDH, succinate dehydrogenase; SFA, saturated fatty acid; suc, succinyl-CoA ligase; TPI, triosephosphate isomerase; UFA, unsaturated fatty acid.