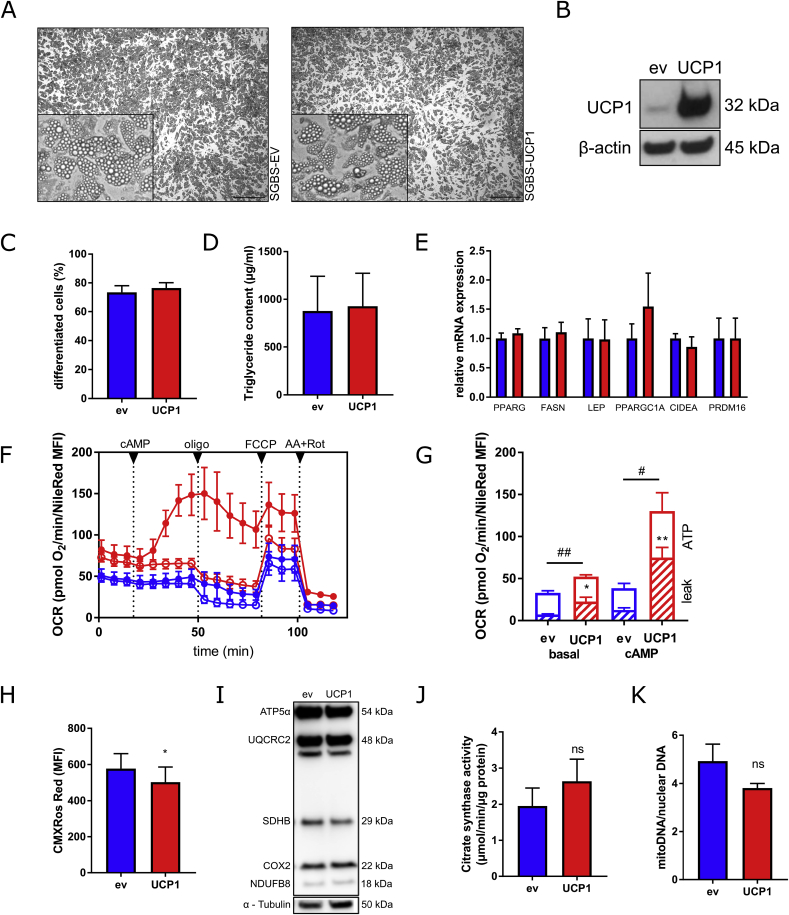
Fig. 4.
Adipogenic differentiation of human preadipocytes overexpressing UCP1. (A) The cells were subjected to adipogenic differentiation for 14 days. (B) UCP1 protein expression was determined by Western Blot. (C) The percentage of differentiated cells was determined by counting preadipocytes and adipocytes using a net micrometer. (D) Lipid accumulation was assessed by measuring the triglyceride content. (E) mRNA expression of key adipogenic marker genes as well as BAT associated genes was determined by qRT-PCR. (F) Oxygen consumption rates (OCR) of UCP1-overexpressing (red color) and control adipocytes (blue color) were determined using plate-based respirometer (media containing 1% BSA). To induce UCP1 activity, subsets of cells were stimulated with 0.5 mM dibutyryl-cAMP (cAMP, closed circles), while control cells were left untreated (open circles) (G) Sequential injections of oligomycin (2 μM), FCCP (4 μM), and antimycin A (1.5 μM) + rotenone (1.5 μM) were used to determine basal, proton leak (striped column part), and ATP-linked respiration rates (open column part). Data were normalized for cellular lipid content by NileRed staining. (H) Mitochondrial membrane potential was measured by CMXRos Mitotracker Red staining. (I) The protein levels of key electron transport chain components were determined by Western Blot. (J and K) To determine mitochondrial content, activity of citrate synthase (J) and relative mitochondrial DNA content (K) were assessed. Data of at least 4 independent experiments are shown as mean + SEM. */#, p < 0.05; **/##, p < 0.01; ns, not significant (Student's t-test). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)