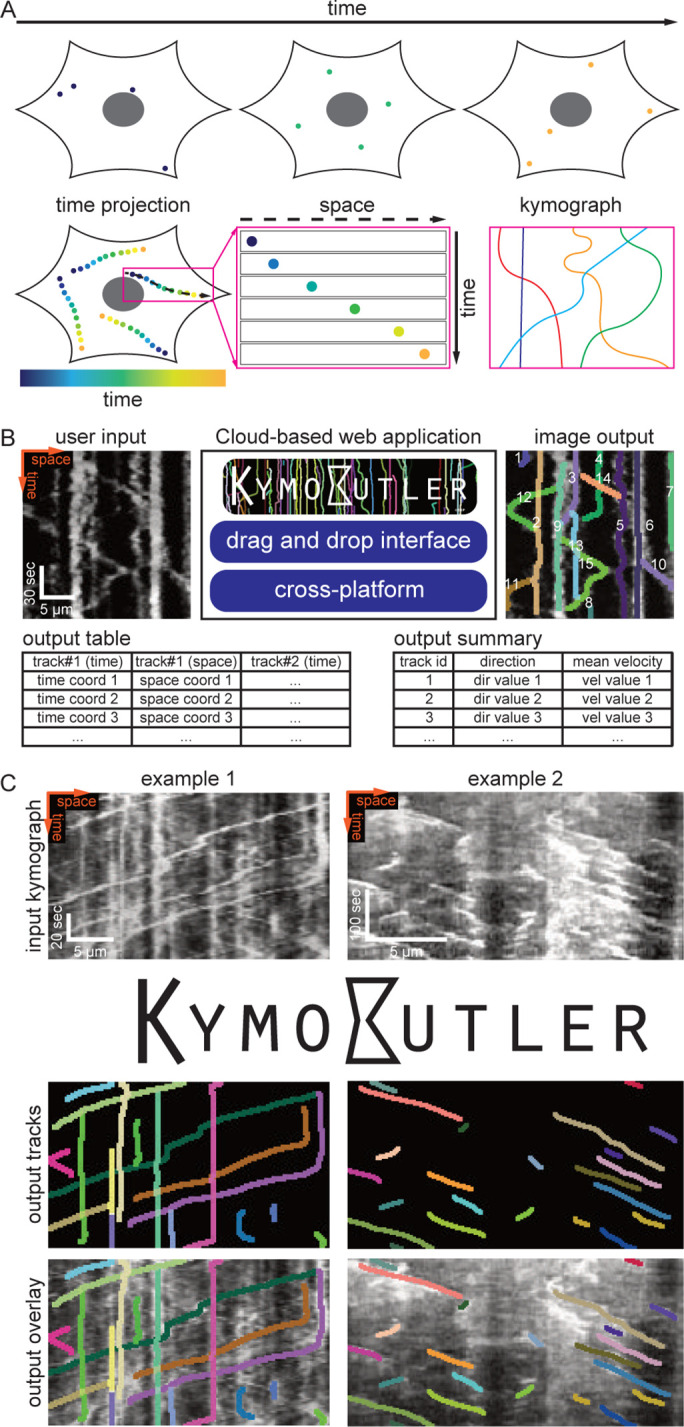
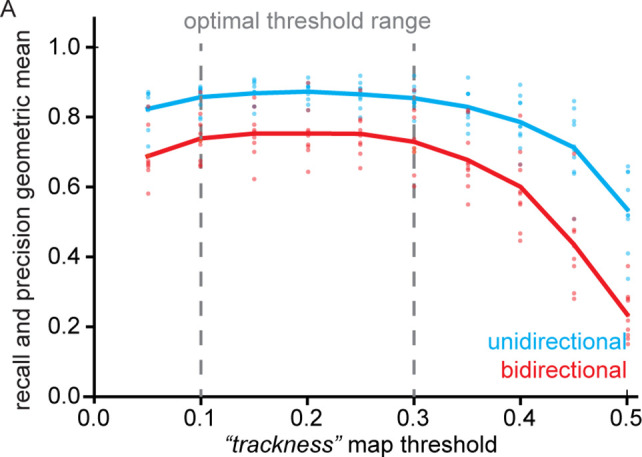
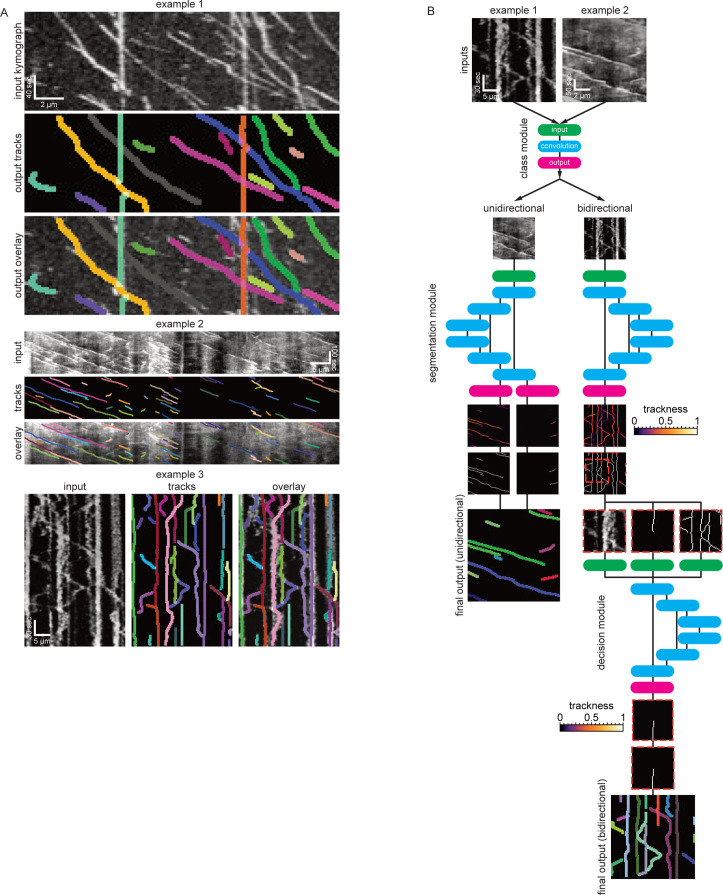
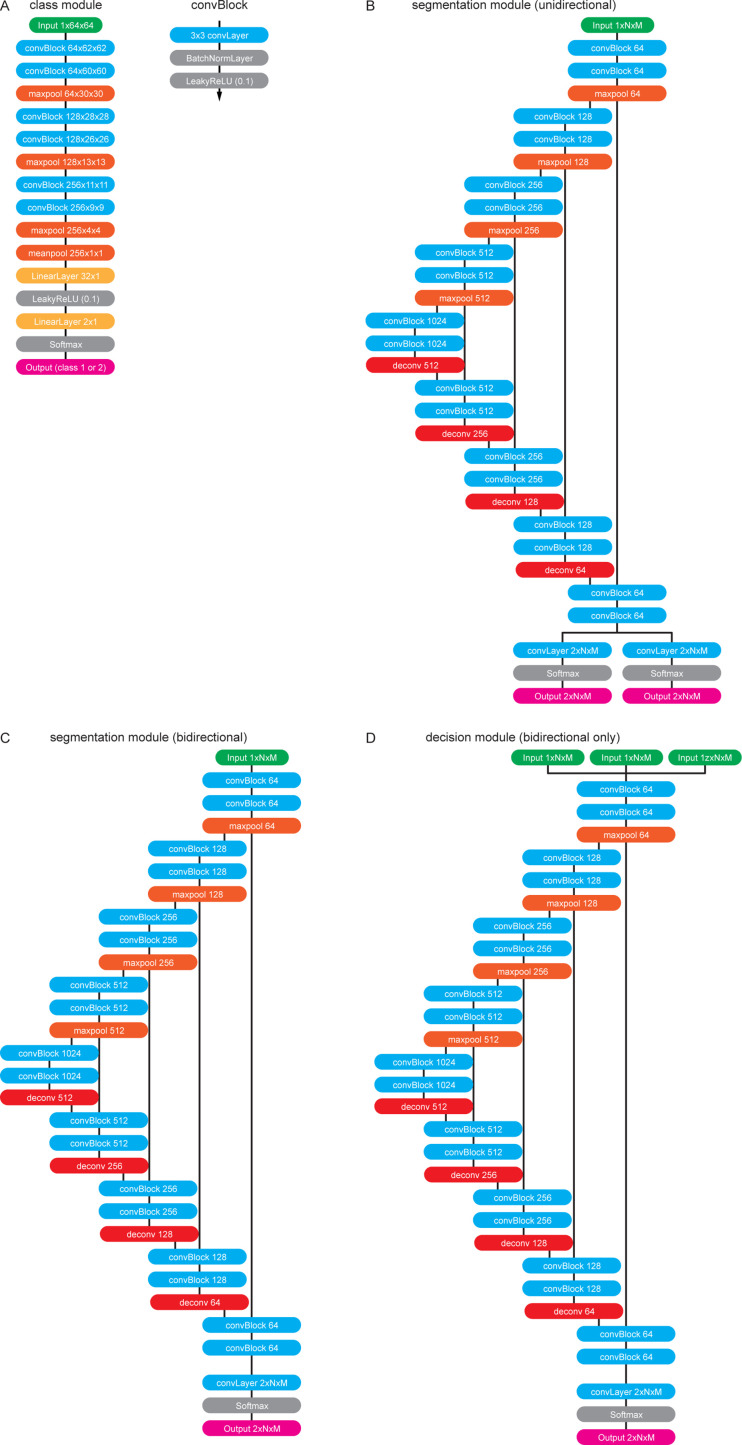
(
A) Three example kymographs from published manuscripts. Example 1: In vitro dynamics of single cytoplasmic dynein proteins adapted from
Tanenbaum et al. (2013). Example 2: EB1-GFP labelled growing microtubule plus-ends in mouse dorsal root ganglion axons (
Lazarus et al., 2013). Example 3: Mitochondria dynamics in mouse retinal ganglion cell dendrites (
Faits et al., 2016). Each dilated coloured line depicts an identified track. (
B) KymoButler software workflow. First, a classification module is applied to each kymograph to determine whether the kymograph is unidirectional or bidirectional. If the kymograph is deemed unidirectional the unidirectional segmentation module is applied to the image to generate two trackness maps that assign each pixel a score between 0–1, approximating the likelihood that this pixel is part of a track with negative slope (left image) or positive slope (right image). Subsequently, the trackness maps are binarized, skeletonised, and segmented into their respective connected components. Finally, those components are averaged over each row to generate individual tracks, and a dilated representation of each track is plotted in a random colour. If the kymograph is classified as bidirectional, another segmentation module is applied to the kymograph, which generates a trackness map that does not highlight any particular slope. This map is binarized with a user-defined threshold and subsequently skeletonised, resulting in a binary map that exhibits multiple track crossings. To resolve these crossings, we first apply a morphological operation that detects the starting points of tracks in the binary map (red dots). Then, the algorithm tracks each line from its starting point until a crossing is encountered. At each crossing, the decision module is called, whose inputs are (i) the raw kymograph in that region, (ii) the previous track skeleton, and (iii) all possible tracks in that region. The decision module then generates another trackness map that assigns high values to the most likely future path from the crossing. This map is then again binarized and thinned with a fixed threshold of 0.5. If the predicted path is longer than two pixels, the path tracking continues. Once all starting points have been tracked until an end (either no prediction or no further pixels available), the algorithm again looks for starting points in the skeletonised trackness map excluding the identified tracks, and repeats the steps outlined above until all pixels are occupied by a track. The resulting tracks are then drawn with each track in a random colour.