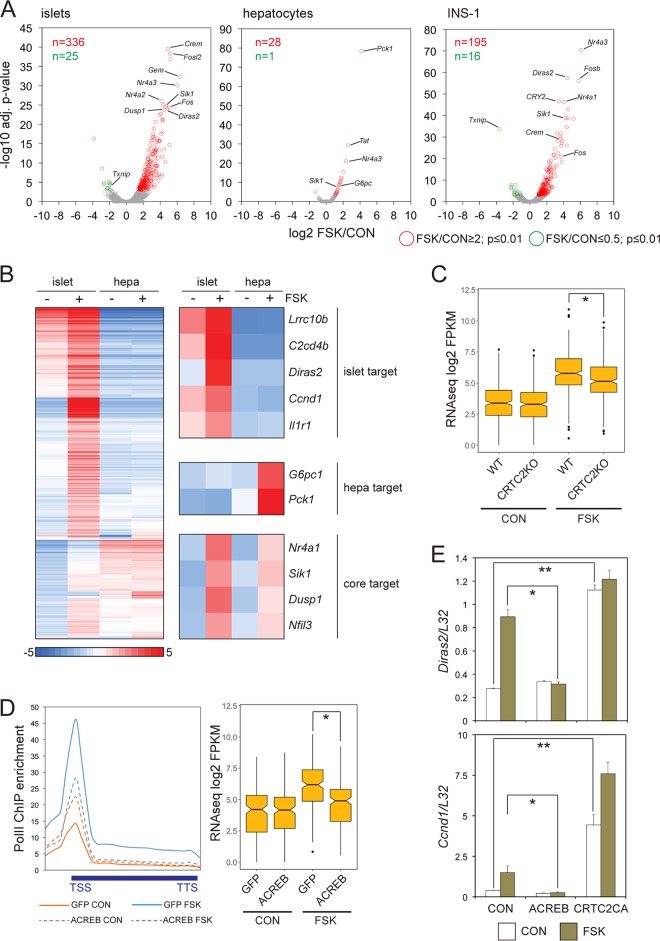
FIG 1.
CREB mediates a broad transcriptional response to cAMP in pancreatic beta cells. (A) Volcano plots of RNA-seq data comparing genomic responses to cAMP in cultured primary mouse islets, primary hepatocytes (hepa), and INS-1 insulinoma cells (n = 2 independent experiments for each; 2 h of FSK exposure). adj., adjusted. (B) Heat map comparing the magnitude of gene induction in response to cAMP in cultured mouse islets relative to primary hepatocytes. Genes upregulated 2-fold or better in primary mouse islets or hepatocytes selected from RNA-seq experiments conducted independently from replicates shown in panel A. Tissue-specific and ubiquitously induced (core) CREB target genes are highlighted. (C) Box plot showing relative effect of FSK on gene expression in cultured pancreatic islets from wild-type and CRTC2 whole-body knockout littermates (n = 882; *, P < 1 × 10−8). (D, left) Metagene analysis of RNA Pol II occupancy over cAMP-induced genes in INS-1 cells infected with control adenovirus (GFP) or adenovirus expressing dominant-negative ACREB. (Right) Effect of ACREB expression on cAMP-induced gene expression relative to that of control (GFP) INS-1 cells. *, P < 1 × 10−11. (E) Effect of adenovirally encoded ACREB inhibitor or phosphorylation-defective, constitutively active CRTC2 [CRTC2CA(S171A)] on expression of beta cell-specific CREB target genes. mRNA amounts for Diras2 and Ccnd1 are shown. *, P < 0.05; **, P < 0.01.