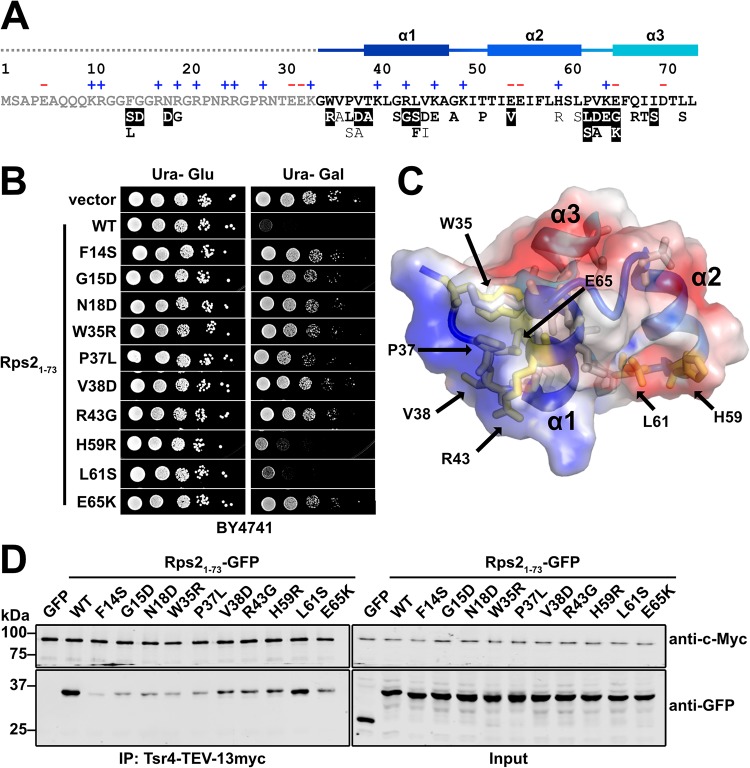
FIG 4.
Tsr4 recognizes two regions of the N terminus of Rps2. (A) Primary sequence of the N-terminal 73 residues of yeast Rps2, with charged residues indicated (+ and −) and secondary structure elements from PDB code 4V88 indicated at the top. The dashed line indicates a region not resolved in the structure under PDB code 4V88. Mutations that alleviated the dominant negative growth phenotype of Rps21–73-GFP are listed below the primary sequence. Strong mutants (white text), moderate mutants (bold black text), and weak mutants (black text) are indicated at the bottom. (B) Growth of a subset of mutants that relieved the dominant negative phenotype of Rps21–73-GFP. Tenfold serial dilutions of wild-type (BY4741) cells expressing the indicated mutants under the control of the galactose-inducible GAL1 promoter were plated on glucose- or galactose-containing medium lacking uracil and grown for 2 days at 30°C. (C) The helical bundle of Rps2 residues 34 to 73, shown as a cartoon depiction with a transparent electrostatic surface (from PDB code 4V88). Positive, negative, and neutral surfaces are indicated by blue, red, and white, respectively. Amino acids that can be mutated to alleviate the growth inhibition of Rps21–73 are shown as sticks, with residues included in panel B shown in yellow. (D) Strength of the interaction between Tsr4 and wild-type (WT) Rps21–73-GFP or the indicated Rps21–73-GFP mutants, as determined by their ability to coimmunoprecipitate with Tsr4-TEV-13myc.