Figure 1.
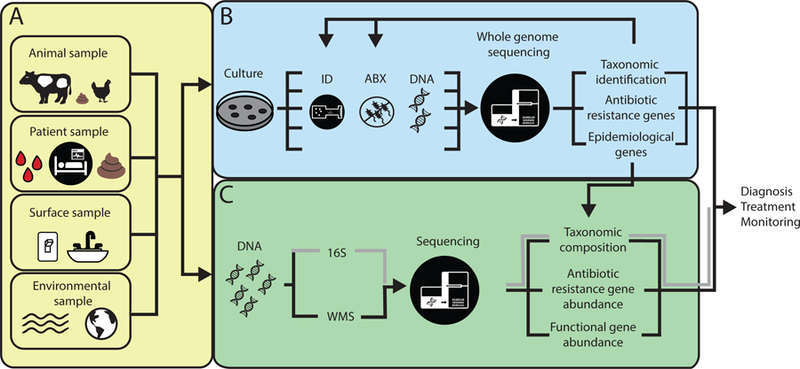
Biological and environmental samples can be interrogated for ARO and ARG enrichment using whole genome sequencing (WGS) and metagenomics analyses. A) Samples can be collected from diverse origin, including biological samples such as tissue, blood, or fecal samples from animal or patient samples. Environmental samples can be collected directly from built environment surfaces or natural environments such as soil and water sources. B) In culture-based methods, isolates are extracted from samples and processed individually to determine species identification and antibiotic susceptibility. After DNA extraction, each isolate can be sequenced using WGS. WGS analyses can determine taxonomic identification, antibiotic resistance genotype, and epidemiological genes. These characteristics can be compared to culture-based results, creating genotype-phenotypic characterization of isolates and improved characterization of species and resistome. WGS analyses can also be used to improve sequence based searches for metagenomics analyses. C) In metagenomics analyses, genomic DNA is extracted directly from a sample, then 16S or whole metagenomics sequencing is done. After 16S amplification and sequencing, taxonomic composition is determined. After WMS, taxonomic composition, antibiotic resistance gene abundance, and functional gene abundance can be analyzed. All of these data can be combined to improve diagnosis, treatment, and ARO and ARG surveillance.
