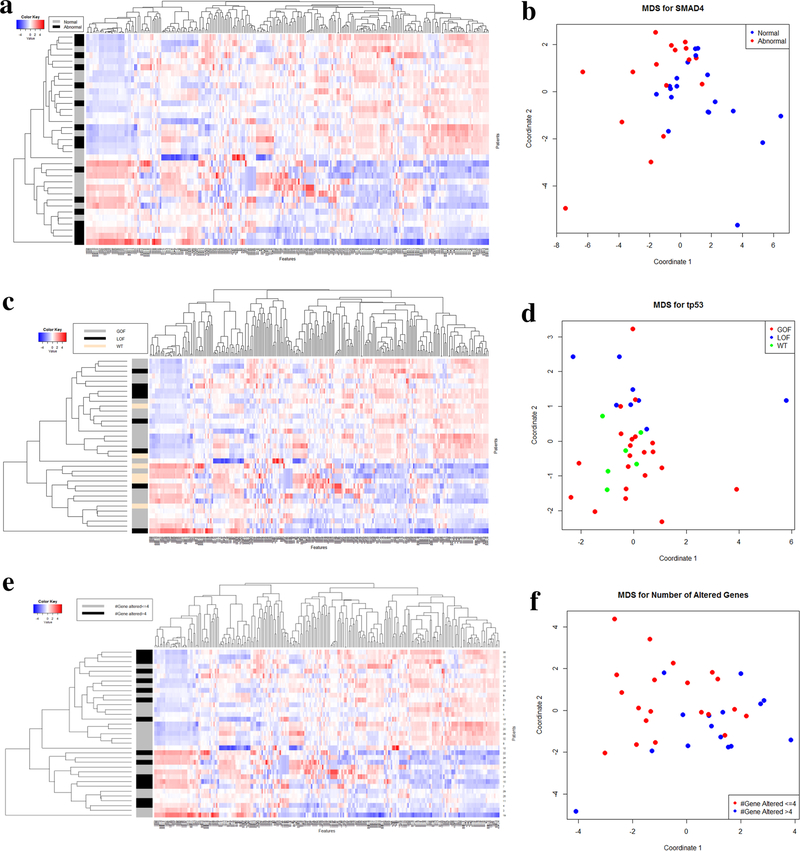
Fig. 3.
Heatmap with classification of 255 texture features and 35 patients for SMAD4 (a), TP53 (c), and number of genes altered (e). Multidimensional scaling for SMAD4 using fMRMR for feature selection (b), TP53 using univariate analysis (p < 0.05) for feature selection (d), and number of genes altered separated by ≤ 4 and > 4 genes altered (f). GOF gain-of-function, LOF loss-of-function, WT wild type