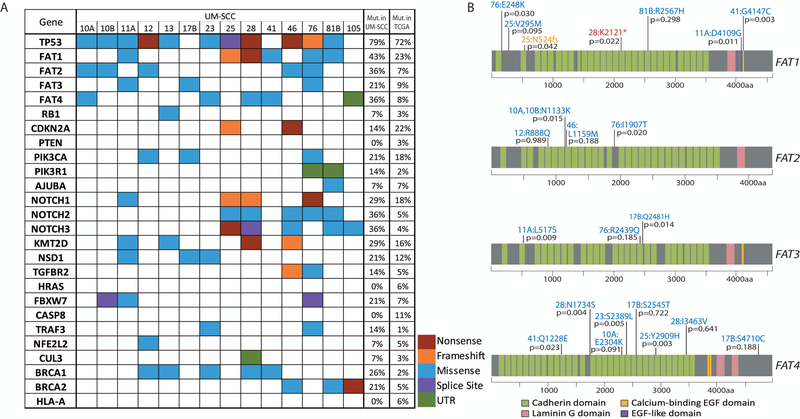
Figure 1. Single nucleotide variants identified in the laryngeal UM-SCC cell line panel.
(A) Non-synonymous mutations in laryngeal UM-SCC cell lines as called using Nimblegen capture-based exome sequencing were color coded by mutation type as indicated. Genes selected for this analysis were previously found to be mutated in the HNSCC TCGA project (5), or were otherwise of interest as common cancer-related genes (FAT, NOTCH, and BRCA families). SNPs occurring in introns were excluded with the exception of those affecting splice sites. Also excluded were SNPs occurring at a frequency >0.01 according to the 1000 Genomes Project. The percentage of cell lines in the LSCC panel harboring a mutation in each gene is indicated immediately to the right. The percentage of TCGA HNSCC samples (TCGA provisional dataset) harboring a point mutation in each gene is noted in the rightmost column (http://cancergenome.nih.gov/). (B) Schematic diagrams of FAT1–4 proteins showing distribution of mutations found in the FAT1 gene in UM-SCC laryngeal cell lines. Resulting amino acid changes are noted for each mutation next to the cell line in which it was identified. VEST-4 pathogenicity score is indicated as a p-value for each mutation.