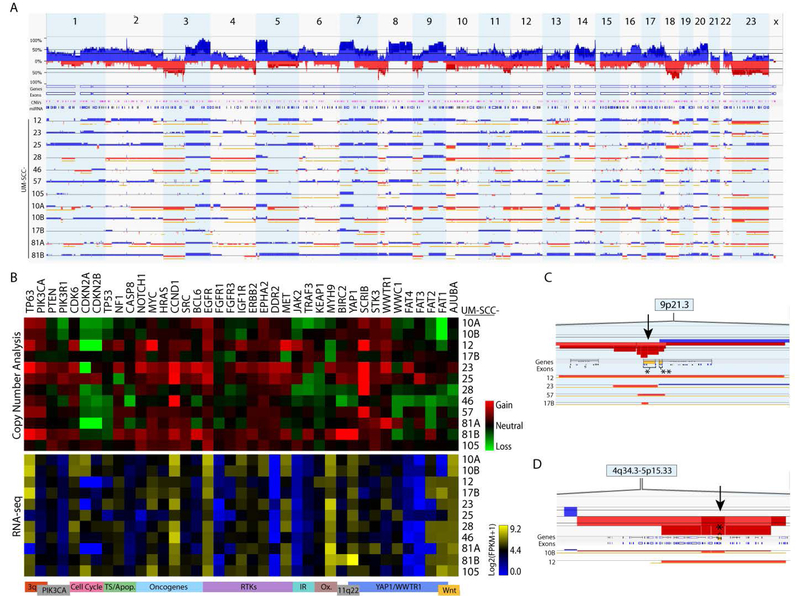
Figure 2. Genetic characterization of laryngeal UM-SCC cell lines by copy number analysis.
Genomic DNA was harvested from low passage UM-SCC cell lines and analyzed using high density SNP arrays (Affymetrix OncoScan Assay) and compared to a commercially available pooled control. Affymetrix software was used to call copy number alterations. (A) Copy number alterations were summed across UM-SCC-10A, 10B, 12, 17B, 23, 25, 28, 46, 57, 76, 81A, 81B, and 105. Alterations for individual cell lines are shown below with gains indicated in blue and losses indicated in red. (B) Heat maps displaying median copy numbers (upper panel) and RNA expression (lower panel) for selected genes. Key functions and relevant chromosomal regions are noted below each column. (C) Focal deletions (arrow) at the CDKN2A-CDKN2B (9p21) locus occurring in UM-SCC 12, 23, 57, and 17B. The CDKN2A gene is indicated by a bracket in the row labeled “Genes.” CDKN2A,* CDKN2B**. (D) Focal deletions (arrow) at the FAT1 locus (4q35) occurring in UM-SCC-10B and 12. The FAT1 locus is indicated by a bracket and an asterisk in the row labeled “Genes.”