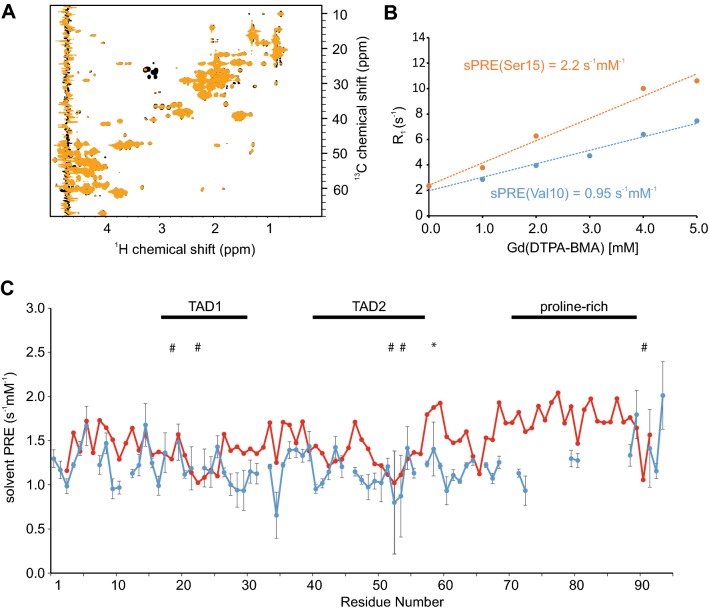
Fig. 3.
Comparison of predicted and measured solvent PRE of p53TAD. a Overlay of 1H, 13C HSQC read-out spectra, with full recovery time of a 300 µM 13C, 15N labeled p53TAD in absence (black) and presence of 5 mM Gd(DTPA-BMA) (orange). b Gd(DTPA-BMA)-concentration-dependent R1 rates of two selected residues. c Diagram showing predicted (red) and measured (blue) solvent PRE values of each Hα atom of p53TAD. Experimental sPRE values are calculated by fitting the data with a linear regression equation. Predicted sPRE values are based on the previously described ensemble approach. Regions binding to co-factors (TAD1, TAD2) and the proline rich region are labeled. Residues with bulky side chains (Phe, Trp, Tyr) are labeled with #, and exposed glycine residues are labeled with * (see Supporting Fig. 2B for a bulkiness profile). Errors of the measured 1H-R1 rates were calculated using a Monte Carlo-type resampling strategy and are shown in the diagram as error bars