Abstract
The excitability of neurons is tightly dependent on their ion channel repertoire. Among these channels, the leak sodium channel NALCN plays a crucial role in the maintenance of the resting membrane potential. Importantly, NALCN mutations lead to complex neurodevelopmental syndromes, including infantile hypotonia with psychomotor retardation and characteristic facies (IHPRF) and congenital contractures of limbs and face, hypotonia and developmental delay (CLIFAHDD), which are recessively and dominantly inherited, respectively. Unfortunately, the biophysical properties of NALCN are still largely unknown to date, as well as the functional consequences of both IHPRF and CLIFAHDD mutations on NALCN current. Here we have set-up the heterologous expression of NALCN in the neuronal cell line NG108-15 to investigate the electrophysiological properties of NALCN carrying representative IHPRF and CLIFAHDD mutations. Several original properties of the wild-type (wt) NALCN current were retrieved: mainly carried by external Na+, blocked by Gd3+, insensitive to TTX and potentiated by low external Ca2+ concentration. However, we found that this current displays a time-dependent inactivation in the −80/−40 mV range of membrane potential, and a non linear current-voltage relationship indicative of voltage sensitivity. Importantly, no detectable current was recorded with the IHPRF missense mutation p.Trp1287Leu (W1287L), while the CLIFAHDD mutants, p.Leu509Ser (L509S) and p.Tyr578Ser (Y578S), showed higher current densities and slower inactivation, compared to wt NALCN current. This study reveals that heterologous expression of NALCN channel can be achieved in the neuronal cell line NG108-15 to study the electrophysiological properties of wt and mutants. From our results, we conclude that IHPRF and CLIFAHDD missense mutations are loss- and gain-of-function variants, respectively.
Subject terms: Channelopathies, Ion channels in the nervous system, Excitability, Membrane potential
Introduction
The maintenance of the neuronal resting membrane potential (RMP) and its modulation by hormones and neurotransmitters is a finely tuned process that involves a complex interplay between several classes of proteins including ion channels, exchangers, transporters, and G protein-coupled receptors1,2. Of importance, Lu and colleagues, in 2007, found that the sodium (Na+)-leak channel NALCN that conducts a background sodium influx critically regulates the RMP of neurons3. Disruption of NALCN expression in mice led to a ≈15 mV hyperpolarization of the RMP of hippocampal neurons and a decrease of their firing rate3. Of note, the homozygous knockout mice died within 24 h after birth as a consequence of a disrupted respiratory rhythm. The importance of NALCN in setting-up the neuronal RMP was then confirmed in other types of neurons and other species, showing a conserved role across species4–11. In addition, NALCN was also found to be involved in the modulation of the neuronal RMP by hormones and neurotransmitters. Indeed, it was demonstrated that NALCN channel could be activated by substance P, neurotensin, and acetylcholine8,11–14. This positive modulation of NALCN involves the corresponding G-protein coupled receptors through a G-protein-independent, src family of tyrosine kinase-dependent pathway12,13. Conversely, a negative modulation of NALCN activity was reported by activation of the the G-protein-coupled calcium-sensing receptor15, the GABA-B receptor16 as well as the Dopamine D2 receptor16. This NALCN inhibition involves a G-protein-dependent pathway15,16.
Both recessive and dominant pathogenic variants of human NALCN and its Unc80 ancillary subunit were recently described in several clinical and genetic studies17–43. Recessive pathogenic variants of NALCN and Unc80 are linked to a syndrome refered to as infantile hypotonia with psychomotor retardation and characteristic facies (IHPRF) type 1 (OMIM #615419) and type 2 (OMIM #616801), respectively. Dominant pathogenic variants of NALCN are responsible for a syndrome refered to as congenital contractures of limbs and face, hypotonia and developmental delay (CLIFAHDD; OMIM #616266). Both IHPRF and CLIFAHDD patients exhibit complex clinical traits of variable severity that may cause premature death in some cases. Interestingly, IHPRF and CLIFAHDD patients exhibit common symptoms such as hypotonia, facial dismorphisms, global developmental delay, constipation, and respiratory defects. Most, but not all, of the recessive IHPRF variants are predicted to result in truncated and non-functional proteins resulting in a loss-of-function phenotype. However, the electrophysiological and functional consequences of the IHPRF mutations are not known to date. Similarly, there is no functional data regarding the CLIFAHDD mutations. Of interest, all the CLIFAHDD mutations are missense and localize in the pore-forming region of NALCN, except the R1181Q mutation that localizes in the intracellular loop linking the transmembrane domains III and IV of NALCN.
Because the functional properties of recombinant NALCN are poorly known, we set-up heterologous expression of human NALCN in a neuron-like environment in order to investigate the electrophysiological properties of the wt, and the IHPRF and CLIFAHDD pathogenic variants of NALCN. We describe here that NALCN currents could be recorded in the neuronal cell line NG108-15 transfected with human NALCN constructs. The overexpressed NALCN current exhibits a time-dependent current decay that resembles inactivation. We also report that the W1287L IHPRF variant resulted in a non-functional NALCN channel, while the L509S and Y578S CLIFAHDD variants promote NALCN currents larger than the wt NALCN current. Importantly, both L509S and Y578S mutations impaired the inactivation kinetics of the NALCN currents, compared to wt NALCN channel. This study provides the first description of the functional consequence of the pathogenic NALCN missense mutations and suggests that loss and gain of NALCN activity supports the inheritance patterns of IHPRF and CLIFAHDD, respectively.
Materials and Methods
DNA constructs
The pcDNA3-GFP, pcDNA3-eGFP-hNALCN-wt, pcDNA3-eGFP-hNALCN-L509S, and the pcDNA3-eGFP-hNALCN-Y578S plasmids were previously described13,21. The pcDNA3-eGFP-hNALCN-W1287L plasmid was generated from the pcDNA3-eGFP-hNALCN-wt plasmid by using a site-directed mutagenesis standard protocol. The W1287L mutation was originally reported in17. The pRK5-mNLF1, pRK5-mNLF1-mCherry, and the pRK5-mNLF1-eGFP plasmids were kind gifts from Drs. M. Zhen and K. Aoyagi.
Cell culture
NG108-15 cells (ATCC® HB-12317™) were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with GlutaMax, 10% dialyzed fetal calf serum, 2% hypoxanthine/aminopterin/thymidine, and penicillin/streptomycin 1% (Invitrogen, Thermo Fisher Scientific). Cells with 70–80% of confluency were transfected in 35 mm culture dishes by using the JetPEI reagent (QBiogen, #101-40N) according to the manufacturer’s procedure. 24 h after transfection, cells were dissociated with Versene (Thermo Fisher Scientific, #15040-033) and plated at a dilution of 1:100 per 35 mm Petri dish for electrophysiological recordings. Differentiation of NG108-15 cells into neuron-like cells was then induced during 5 days by replacing the culture media by the same medium with lowered serum concentration to 1% and the addition of dbcAMP 200 µM (Sigma-Aldrich, #D0627) and dexamethasone 0.1 µM (Sigma-Aldrich, #D4902). Analysis of NALCN expression (PCR, Western-blot and patch-clamp experiments) was performed 6 to 7 days post-transfection. The control condition (Mock) corresponds to cells transfected with NLF1 alone.
Non-quantitative (nqPCR) and quantitative PCR (qPCR)
Total RNA was extracted from cell cultures and rat tissues using the RNeasy Plus universal mini kit (Qiagen #74134). One μg of total RNA were reverse transcribed using the iScript kit (BioRad #1708891) according to the manufacturer’s protocol. For nqPCR, Taq polymerase (New England Biolabs #M0273S) and dNTP (New England Biolabs #N0447S) were used according to the manufacturer’s protocol with the following parameters: 94 °C-3min, 30 cycles of 94 °C-45sec, 55 °C-45sec, 68 °C-90sec, and a final step at 68 °C-10min. Primers used for nqPCR were: NALCN-sense 5′-TTCTTTGTTGGATGCCTCAAA-3′, NALCN-antisense 5′-GGCCCCACACGATGAATAAT-3′, Unc79-sense 5′-TTGGATTTCCAGAGCAATCAA-3′, Unc79-antisense 5′-ACCTGGGCCATTTTGAACTG-3′, Unc80-sense 5′-AAGCTGGCACCATATGACAC-3′, Unc80-antisense 5′-ACAACAGCTGTGCTGAAGGG-3′, NLF1-sense 5′-GGCTCTTTTTCTAGGAAACTC-3′, NLF1-antisense 5′-CACGCTTTCAAACTCTTCGT-3′, APP-sense 5′-CAGGTCATGAGAGAATGGGA-3′, APP-antisense 5′-ACGGAGGTGTGTCATAACCT-3′. For qPCR, RT products were diluted 10 times with H2O and stored at −20 °C until use. Real-time PCR was performed in 384 well plates in a final volume of 10 μl using SYBR Green dye detection on the LightCycler480 system (Roche-Diagnostic #04887352001). The primer pairs were designed using Primer 3 input software, and their specificity and efficacy was experimentally validated. Cq for individual genes were determined using the second Derivative Max tool of the LightCycler480 software. The relative RNA expression was calculated using the RNA helicase Ddx17 as a reference gene. Relative expressions were calculated using the ΔΔCq method44. Primers used in this study were: NALCN-sense 5′-ccagagctagaagaaggctatca-3′, NALCN-antisense 5′-tgcctgctaagtcccaggt-3′, Unc80-sense 5′-ctctgtggatcgcctgtctt-3′, Unc80-antisense 5′-gacagcgctggtgaacttg-3′, Unc79-sense 5′-gccgaggaatcagaatttaaga-3′, Unc79-antisense 5′-gctgacggcaatcttcctt-3′, NLF1-sense 5′-aaccgaggcaagaacaacc-3′, NLF1-antisense 5′-gtctctaggcgccacacc-3′, Ddx17-sense 5′-aagttgatgcagcttgtgga-3′, Ddx17-antisense 5′-gaagtagtccggtatcgtgagc-3′.
Biotinylation of proteins at the cell surface
Cells were washed twice with cold Phosphate Buffered Saline (PBS, Life Technologies #14040-091). For biotinylation, the cells were incubated in cold biotinylation solution (0.5 mg/ml EZ-Link Sulfo-NHS-SS-Biotin, Thermo Fisher Scientific #21331 in PBS) for 30 min on ice. The biotinylation solution was removed, and the cells were washed three times with stop solution (10 mM Tris-HCl, pH 7.4, 120 mM NaCl). Cells were lysed in lysis buffer (10 mM Tris-HCl, pH 7.4, 120 mM NaCl, 1% Triton-X-100 (v/v)) supplemented with proteases inhibitors (Roche Applied Science #04693124001) and centrifuged at 10,000 × g for 30 min at 4 °C. The clear supernatant was incubated with NeutrAvidin agarose (Thermo Fisher Scientific #29200) for 2 hours at 4 °C. After incubation, the beads were washed three times with lysis buffer and proteins were eluted by adding laemmli buffer then submitted to immunoblotting.
Immunoblotting
Cells were lysed on ice for 20 min with NP-40 buffer containing 10 mM Tris-HCl, pH 7.4, 120 mM NaCl, 1% NP-40, and proteases inhibitors (Roche Applied Science #04693124001). Cell lysates were spun at 10,000 × g for 30 min at 4 °C. Sixty micrograms of proteins were mixed with a 4x Laemmli buffer and then loaded on SDS-PAGE. Proteins resolved onto 4–20% polyacrylamide gels (Bio-rad #4561096) were transferred to Hybond C nitrocellulose membranes (GE Healthcare #10600016). Membranes were immunoblotted with primary antibodies (rabbit anti-GFP, 1:10000, Torrey Pines Biolabs #TP401; rabbit anti-α-actinin, clone D6F6, 1:3000, Cell Signaling #6487; mouse anti-Na+/K+-ATPase, 1:5000, Abcam #AB7671; rabbit anti-calreticulin, 1:1000, Abcam #92516) and then with either an anti-rabbit or anti-mouse horseradish peroxidase (HRP)–conjugated secondary antibody (anti-rabbit, 1:10000, GE Healthcare #NA934; anti-mouse, 1:5000, Merck #12-349). Immuno-reactivity was detected with an enhanced chemiluminescence substrate for detection of HRP (SuperSignal West Pico Chemiluminescent Substrate, Thermo Fisher Scientific #34080).
Electrophysiological recordings
Macroscopic currents were recorded at room temperature using an Axopatch 200B amplifier (Molecular Devices). Borosilicate glass pipettes had a resistance of 3 to 5 MΩ when filled with an internal solution containing (in mM): 150 CsCl, 5 HEPES, 2 MgCl2, 1.1 EGTA (pH adjusted to 7.34 with NaOH, ~300 mOsm). The extracellular solution contained (in mM): 142.6 NaCl, 2 CaCl2, 5.6 KCl, 0.8 MgCl2, and 10 HEPES, 5 Glucose (pH adjusted to 7.35 with NaOH, ~310 mOsm). In some experiments, when indicated, the CaCl2 concentration was reduced to 0.1 mM. The recording chamber was constantly perfused (~100 µl/min) with the control (Na+-containing) solution using a gravity-driven homemade perfusion device, which allowed extracellular medium change and drug application.
Data analysis
Data were analyzed using pCLAMP9 (Molecular Devices) and GraphPad Prism (GraphPad) softwares. Results are presented as the mean ± SEM, and n is the number of cells. Statistical analysis was performed either with the Student t-test or a Mann-Withney test or with one-way ANOVA combined with a Tukey post-test for multiple comparisons (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
Results
The NG108-15 cell line endogenously expresses Unc79 and Unc80
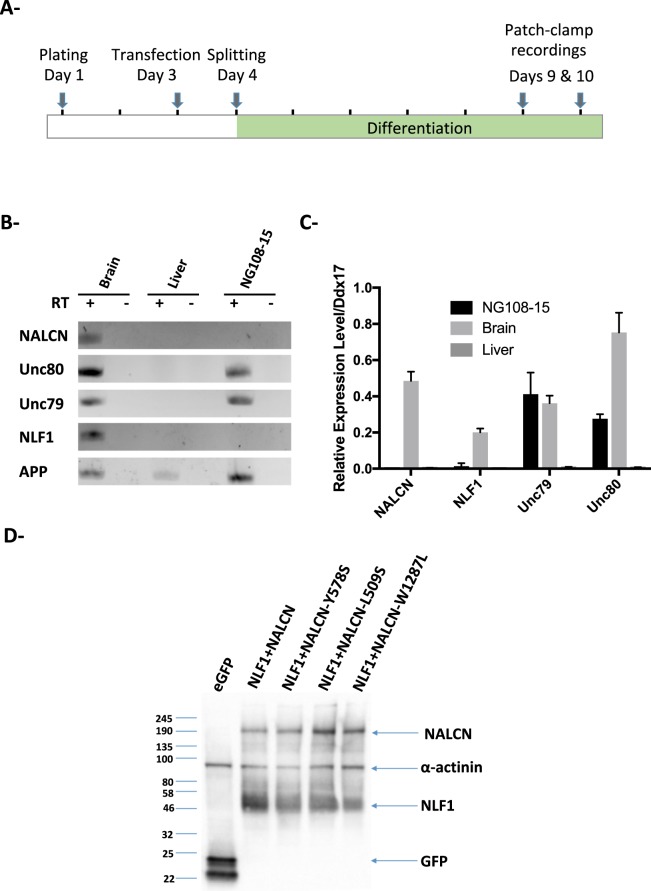
We previously demonstrated that the NG108-15 cell line is a suitable neuronal model for the study of voltage-gated calcium channels45,46. Importantly, NG108-15 cells can be differentiated into cholinergic neuron-like cells by adding dbcAMP and dexamethasone for 4–5 days (see the Materials and Methods section; Fig. 1A)47. In order to determine whether differentiated NG108-15 cells could be used to functionally express NALCN, we checked for the expression of the known components of the NALCN channelosome by RT-nqPCR and RT-qPCR (Fig. 1B,C). As a matter of fact, NALCN belongs to a large protein complex that includes, in addition to NALCN, at least 3 additional subunits: Unc79, Unc80 and NLF1 (reviewed in48). Both the Unc79- and the Unc80-encoding mRNAs were detected in differentiated NG108-15 cells, but not NALCN nor NLF1. However, because of the lack of commercially available antibodies, we were not able to confirm protein expression by Western blotting. We next examined by Western blot whether missing components of the NALCN channelosome (e.g. NALCN and NLF1) could be provided by transfection of ectopic cDNAs. We found that wt NALCN, and the pathogenic variants, as well as and NLF1 were expressed at high level in transfected NG108-15 cells 6 days after transfection and differentiation (Fig. 1D).
Figure 1.
The NG108-15 cell line endogeneously expresses Unc79 and Unc80. (A) Schematic representation of the protocol used to achieve differentiation of NG108-15 cells into neuron-like cells. (B) RT-nqPCR with total RNA extracted from differentiated NG108-15 cells, rat adult brain and rat adult liver. The + and − symbols indicates the presence or the absence of the Reverse Transcriptase respectively. The ubiquitously expressed transmembrane Amyloid Precuror Protein (App)-encoding mRNA was used as an internal control. (C) RT-qPCR results with the same RNA samples than in B- showing that both Unc79 and Unc80, but not Nalcn and Nlf1, are expressed in differentiated NG108-15 cells. The gene encoding the RNA helicase Ddx17 was used as a housekeeping gene (n = 4). (D) Representative western blot showing that transfection of the wt NALCN- and its pathogenic variants W1287L, Y578S, L509S, and NLF1-encoding cDNAs in NG108-15 cells results in a high expression level of the corresponding proteins, but not in the eGFP condition.
Co-expression of NALCN and NLF1 in NG108-15 cells leads to the recording of a Na+ background current
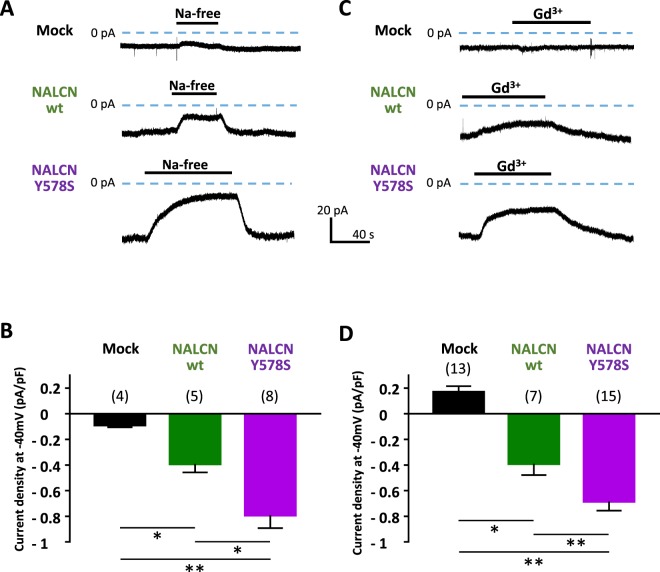
Using the patch-clamp technique, whole-cell recordings were carried out in NG108-15 cells co-transfected with NALCN and NLF1. We first performed experiments to examine the effect of external Na+ removal (NMDG-substituted) when cells were held at −40mV. Of note, we did not observed any effect of Na+ removal on non-differentiated NG108-15 cells (data not shown). Replacing external Na+ by the impermeant cation NMDG in differentiated cells resulted in a significant decrease of a background current in cells co-transfected with NALCN and NLF1 (−0.391±0.14 pA/pF, n = 5), compared to the control condition (Mock; −0.088±0.03 pA/pF, n = 4, p = 0,031, Fig. 2A,B). Importantly, NMDG application revealed that this Na+ background current was even larger in cells expressing the CLIFAHDD variant NALCN-Y578S (−0.79±0.32 pA/pF, n = 8, p = 0.004) and was significant larger than wt NALCN (p = 0.045).
Figure 2.
Transient expression of the wild-type (wt) NALCN and its CLIFAHDD variant Y578S together with NLF1 in differentiated NG108-15 cells results in the expression of a NMDG- and Gd3+-sensitive background Na+ current. (A) Representative traces obtained on cells held at −40 mV (HP) during application/wash of an extracellular Na+-free solution (replaced with NMDG) for a mock (NLF1)-transfected cell (top), a NALCN/NLF1 (wt)-transfected cell (middle), a NALCN CLIFAHDD mutant (Y578S)/NLF1-transfected cell (bottom). (B) Density of the external Na+-dependent background current (Na+-free) for mock-transfected cells (−0.088 ± 0.03 pA/pF, n = 4); NALCN: −0.391 ± 0.14 pA/pF, n = 5; NALCN-Y578S: −0.79 ± 0.32 pA/pF, n = 8). (C) Representative traces obtained on cells held at −40 mV during application/wash of Gd3+ (10 µM) for a mock-transfected cell (top), a NALCN (wt)-transfected cell (middle), a NALCN CLIFAHDD mutant (Y578S)-transfected cell (bottom). (D) Average density of the Gd3+-sensitive background current for mock-transfected cells (0.168 ± 0.16 pA/pF, n = 13), NALCN (wt)-transfected cells (−0,391 ± 0.23 pA/pF, n = 7) and NALCN-Y578S-transfected cells (−0.684 ± 0.27 pA/pF, n = 15). Both the Na+-dependent background current and the Gd3+-sensitive background current were significantly of higher density for wt NALCN and its pathogenic variant Y758S compared to the control condition (p = 0,03 and 0,004 for the Na+-dependent component and P < 0.0001 and <0.0001 for the Gd3+-sensitive component respectively). In addition, densities of the Na+-dependent background current and the Gd3+-sensitive background current were significantly lower for wt NALCN compared to its pathogenic variant Y578S (p = 0.045 and p = 0.029 respectively). p values were calculated with a Mann-Whitney statistical test.
NALCN current was previously reported to be Gd3+-sensitive and TTX-resistant3,12,13. We therefore studied the effect of Gd3+ and TTX on the background current in NALCN-transfected NG108-15 cells. We found that the background current in cells expressing wt NALCN and its pathogenic variant Y578S was inhibited by Gd3+ (10 μM; Fig. 2C,D). In addition, we found no effect of TTX (10 μM) on the background Na+ current in NALCN-transfected NG108-15 cells (data not shown). Taken together, these data indicate that expression of NALCN channel results, in differentiated NG108-15 cells, in the functional expression of a Na+ background current with pharmacological properties reminiscent to those described previously for NALCN currents3,12,13.
The Na+ background current amplitude is differentially affected by the CLIFAHDD and IHPRF1 variants of NALCN
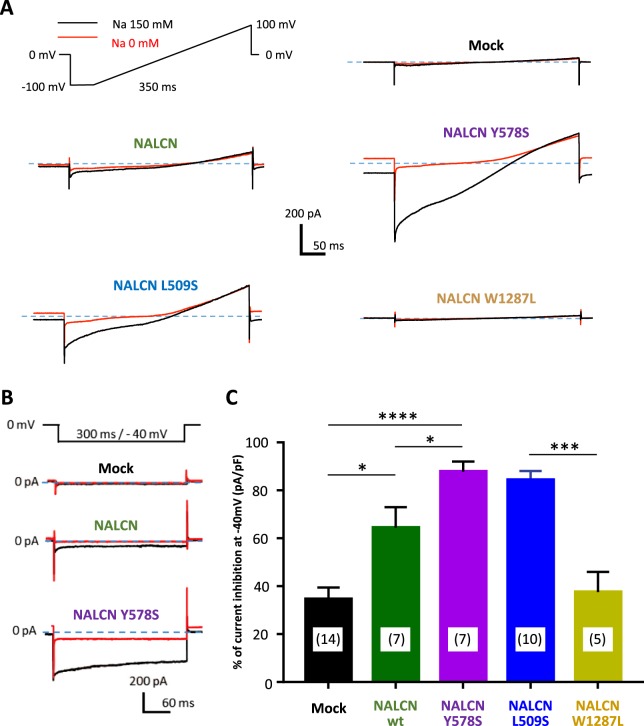
The Na+-dependence of the background current in NALCN-transfected NG108-15 cells was further investigated using voltage ramp and voltage step protocols (Fig. 3). Removal of external Na+ revealed that the CLIFAHDD variants Y578S and L509S conducted a higher Na+ background current compared to wt NALCN. Using a voltage ramp command from −100 mV to +100 mV (Fig. 3A), we observed that the inward Na+ background current was rather linear in a wide range of membrane potentials (−100 to +100 mV) for both wt NALCN and its CLIFAHDD variants. Importantly, no detectable inward Na+ background current was recorded in cells mock-transfected (NLF1 alone) or co-transfected with the IHPRF1 W1287L NALCN mutant. Amplitude of the inward Na+ background current was also measured using one membrane voltage step from 0 mV to −40 mV (Fig. 3B) and normalized by calculating, for each cell, the percentage of inhibition of this current due to the removal of the external Na+ (Fig. 3C). The small current recorded in mock- and the IHPRF1 mutant-transfected cells was weakly reduced in the absence of external Na+ (34.52 ± 18.37%, n = 15, and 37.56 ± 18.70%, n = 5, respectively). Reduction of the Na+ background current was higher in wt NALCN- and the two CLIFAHDD missense mutants (Y578S; L509S)-transfected cells: 64.41 ± 22.67%, n = 7, p = 0.026 for wt, 87.81 ± 12.06%, n = 7, p < 0.0001, for Y578S and 84.25 ± 12.06% n = 10, p < 0.0001, for L509S, NALCN channels. These data clearly indicate that wt NALCN channel generates Na+ background current in the neuronal NG108-15 cells. This Na+ background current is significantly larger for the two CLIFAHDD mutants investigated here, especially the Y578S variant. On the contrary, no Na+ background current could be detected in cells expressing NALCN channels carrying the IHPRF1 missense variant W1287L (p = 0.864).
Figure 3.
Amplitude of the Na+-dependent component of the NALCN current carried out by the CLIFAHDD variants (Y578S, L509S) and the IHPRF variant (W1287L) of the NALCN channel. (A) Representative current traces elicited by a voltage ramp protocol (from −100 mV to +100 mV) in the presence (black traces) and in the absence (red traces) of extracellular Na+. (B) Representative current traces elicited by a voltage step protocol (HP 0 mV, test pulse (TP) at −40 mV) in the presence (black traces) and in the absence (red traces) of extracellular Na+. (C) Percentage of current inhibition obtained in Na+-free condition for mock-transfected cells (34.51 ± 18.37%, n = 14), for NALCN-transfected cells (64.41 ± 22.67%, n = 7), for NALCN-Y578S-transfected cells (87.81 ± 11.2%, n = 7), NALCN-L509S-transfected cells (84.25 ± 12.06%, n = 10) and for NALCN-W1287L-transfected cells (37.56 ± 12.06%, n = 5). Compared to the control condition, the percentage of inhibition was significantly larger for NALCN-wt (p = 0.026), NALCN-Y578S (p < 0.0001), NALCN-L509S (p < 0.0001) but not for NALCN-W1287L (p = 0.864). A significant difference in the percentage of inhibition was also found for NALCN-wt compared to its pathogenic variant Y578S (p = 0.017) but not for L509S (p = 0.157) and W1287L (p = 0.197). P values were calculated with a Mann-Whitney statistical test.
Current-Voltage relationship of wt and mutant Y578S NALCN currents in differentiated NG108-15 cells
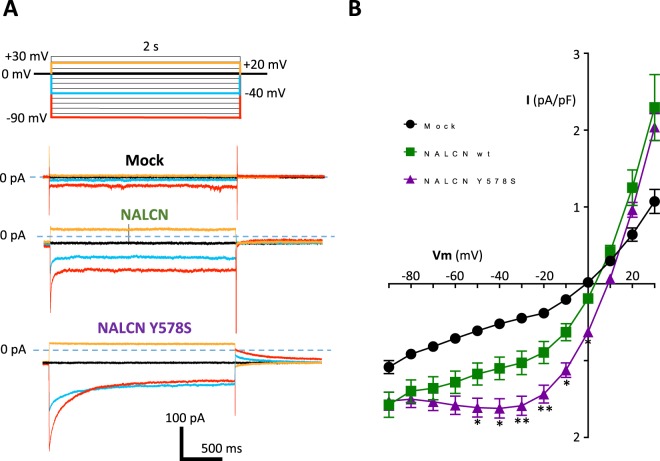
A current-voltage (I/V) step protocol was used to characterize further the electrophysiological properties of the wt and Y578S CLIFAHDD mutant NALCN channels (Fig. 4). Using hyperpolarizing steps from a holding potential (HP) 0 mV, we found that both wt and Y578S NALCN currents displayed a time-dependent inactivation-like decay (Fig. 4A). In addition, our measurements of the steady-state NALCN current (at 2 s) revealed a non-linear I/V relationship for both wt and Y578S NALCN current in NG108-15 cells (Fig. 4B). Similar findings were made with the mutant NALCN L509S (data not shown). Altogether, here we describe that NALCN channels heterogously expressed in differentiated NG108-15 cells exhibit electrophysiological properties indicative of a voltage sensitivity. This finding is novel, considering previous studies that reported a linear, ohmic-like, I/V relationship for the NALCN current, as well as a voltage-independent time-course of the NALCN current3,4,15,32,49–51.
Figure 4.
Current-voltage (I/V) relationship of the steady-state NALCN current for wild-type (wt) and its CLIFAHDD variant Y578S in differentiated NG108-15 cells. (A) Representative current traces elicited by a membrane voltage step protocol (test pulses from −90 mV to + 30 mV) from a HP at 0 mV, for a mock-transfected cell (top), a NALCN (wt)-transfected cell (middle), a NALCN (Y578S)-transfected cell (bottom). Only the current traces obtained for the test-pulses at −90 mV (in red), −40 mV (in blue), 0 mV (in black) and +20 mV (in orange) are shown. (B) Corresponding I/V relationships for mock transfected cells (black circles, n = 44), NALCN wt (green squares, n = 45) and its pathogenic variant NALCN Y578S (purple triangles, n = 73). Statistical significance was calculated with a Tukey’s multiple comparisons test.
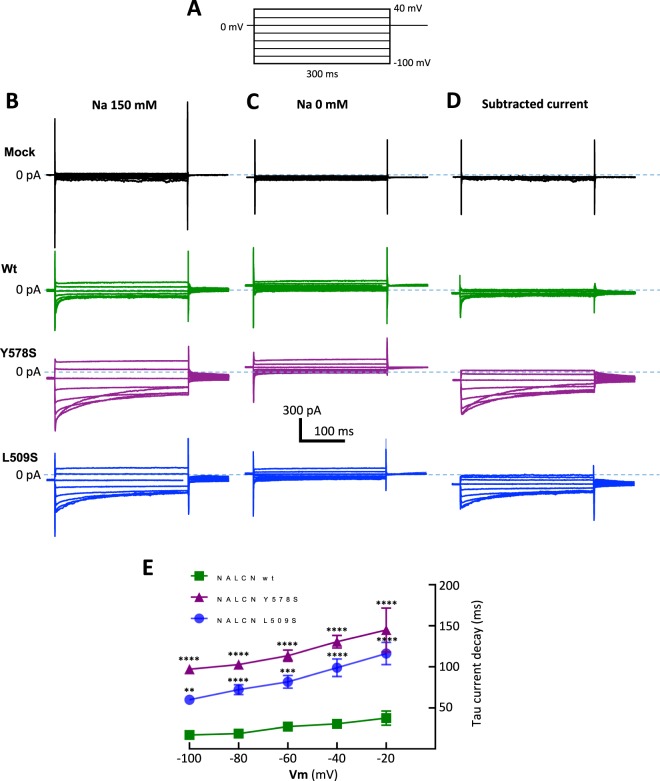
Voltage-dependent inactivation of the NALCN current in differentiated NG108-15 cells
To determine whether the inactivation-like process was readily supported by NALCN channel activity, we examined its dependence to the external Na+ concentration (Fig. 5). The Na+-dependent component (subtracted current, Fig. 5D) of wt NALCN current (green traces), the L509S mutant (blue traces) and the Y578S mutant (purple traces) was isolated by substracting current traces obtained in the presence of extracellular Na+ (Fig. 5B) to the current traces obtained when Na+ was replaced with NMDG (Fig. 5C). Both for the wt NALCN and the two CLIFAHDD mutants, the time-dependent decay of the Na+-dependent component of the NALCN current displayed an inactivation-like component in the −80 mV/−20 mV range of the membrane potential that was best fitted with a single exponential time-constant (Fig. 5E). Of note, kinetics of this time-dependent decay at −40 mV was 3 and 4 times slower, respectively, for the L509S mutant (98.84 ± 10.80 ms) and for the Y578S mutant (130.65 ± 7.65 ms), compared to that measured for the wt NALCN current (30.66 ± 3.60 ms).
Figure 5.
Time- and voltage-dependent inactivation of the NALCN current elicited by the wt, Y578S and L509S variants. (A) Voltage step protocol used to elicit representative current traces obtained in the presence of extracellular Na+ (B), in the absence of extracellular Na+ (C) and the corresponding trace subtraction to isolate the Na+-dependent component of the NALCN current (D) for mock-transfected cells (black traces), NALCN-wt-transfected cells (green traces), NALCN-Y578S-transfected cells (purple traces) and NALCN-L509S-transfected cells (blue traces). (E) Inactivation kinetics (time-constant, τ) of the Na+-dependent component of the wt (n = 7), NALCN-Y578S (n = 7) and NALCN-L509S (n = 6) currents in the −100 mV to −20 mV range. Statistical significance was calculated with a Tukey’s multiple comparisons test.
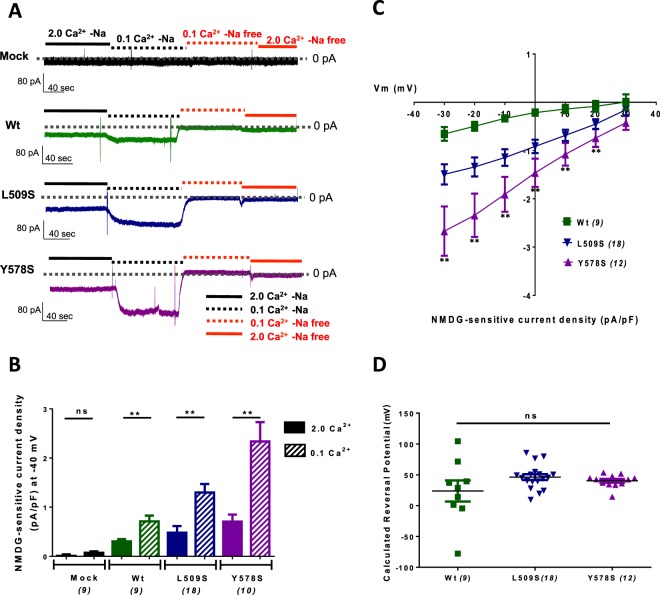
No change in sodium selectivity for NALCN current potentiated in low extracellular Ca2+ concentration
An interesting feature of the NALCN current is its sentivity to extracellular Ca2+. Indeed, a decrease of extracellular Ca2+ from 2 mM to 0.1 mM resulted in a strong enhancement of the NALCN current in native neurons15,16. Therefore, we chose to explore the sensitivity to extracellular Ca2+ of the wt, Y578S and L509S NALCN channels expressed in NG108-15 cells (Fig. 6). As illustrated in Fig. 6A, the reduction of extracellular Ca2+ resulted in a marked increase in the Na+-background current in cells expressing the wt, Y578S and L509S NALCN channels, but not in the control (mock) condition. In average, the NALCN current density was 2 to 3 fold higher in these three conditions when the extracellular Ca2+ concentration was reduced from 2 mM to 0.1 mM (Fig. 6B). We next examined the −30 to +30 mV range of the I/V relationship of the NMDG-sensitive current in cells expressing either wt, L509S or Y578S channels (Fig. 6C) in order to estimate the reversal potential of the corresponding currents. The reversal potential was calculated from the I/V curve of each cell. The mean values of the reversal potentials of the NMDG-sensitive current for wt, Y578S and L509S NALCN channels showed no significant difference (wt NALCN, Erev = 36,55 ± 12,91 mV, n = 8; L509S NALCN, Erev = 46.22 ± 4.70 mV, n = 18; Y578S NALCN, Erev = 40.22 ± 2.99 mV, n = 12; Fig. 6D). These findings indicate that the selectivity to Na+ of NALCN channel was not significantly altered by the L509S and Y78S mutations.
Figure 6.
The NALCN current for wt, Y578S and L509S variants is potentiated in low extracellular Ca2+ with no change in reversal potential. (A) Representative traces obtained on cells held at −40 mV (HP) during application/wash of an extracellular Na+-free solution (replaced with NMDG) with high (2 mM) or low (0.1 mM) extracellular Ca2+ for a mock (NLF1)-transfected cell, a NALCN/NLF1 (wt)-transfected cell, a NALCN CLIFAHDD mutant (L509S)/NLF1-transfected cell, and a NALCN CLIFAHDD mutant (Y578S)/NLF1-transfected cell. (B) Density of the external Na+-dependent background current (Na+-free) with high (2 mM, filled bars) or low (0.1 mM, hatched bars) extracellular Ca2+ for mock (NLF1)-transfected cells (−0.008 ± 0.029 pA/pF in [Ca2+]e = 2 mM and −0.073 ± 0.028 pA/pF in [Ca2+]e = 0.1 mM, n = 9, P = 0.136), NALCN/NLF1 (wt)-transfected cells (−0.303 ± 0.046 pA/pF in [Ca2+]e = 2 mM and −0.712 ± 0.115 pA/pF in [Ca2+]e = 0.1 mM, n = 9, p = 0.004). NALCN CLIFAHDD mutant (L509S)/NLF1-transfected cells (−0.529 ± 0.148 pA/pF in [Ca2+]e = 2 mM and −1.347 ± 0.188 pA/pF in [Ca2+]e = 0.1 mM, n = 16, P = 0.001), and NALCN CLIFAHDD mutant (Y578S)/NLF1-transfected cells (−0.792 ± 0.126 pA/pF in [Ca2+]e = 2 mM and −2.466 ± 0.415 pA/pF in [Ca2+]e = 0.1 mM, n = 16, P = 0.001). Statistical significance was calculated with an unpaired student’s t test (C) I/V relationship in the −30 mV to + 30 mV range of the Na+-dependent component recorded in low concentration of extracellular Ca2+ (0.1 mM) of wt NALCN (n = 8), NALCN-Y578S (n = 12) and NALCN-L509S (n = 18) currents. (D) Mean reversal potential (ERev) of the Na+-dependent component of wt NALCN (Erev = 36.55 ± 12.91 mV, n = 8), NALCN-Y578S (Erev = 40.22 ± 2.99 mV, n = 12) and NALCN-L509S (Erev = 46.22 ± 4.70 mV, n = 18) currents. Each dot represents the ERev of 1 cell. For each cell, the ERev was calculated from the linear part of its I/V relationship as illustrated in (C). Statistical significance was calculated with a Tukey’s multiple comparisons test.
A difference in membrane expression does not appear to account for the gain-of-function effect of L509S and Y578S pathogenic variants
We next performed the cell biotinylation of surface proteins in order to determine whether the gain-of-function effect of the L509S and Y578S NALCN mutants could result from an increase in plasma membrane expression (Fig. 7). Six independent experiments were conducted and a representative one is presented in Fig. 7A. We found that both the total expression and the membrane expression of the L509S and Y578S NALCN mutants (normalized with the Na+/K+ ATPase expression level) were significantly lower than expression of wt NALCN. In average (n = 6), the total expression normalized to wt NALCN was 62.99 ± 8.72% for L509S (p < 0.01) and 56.24 ± 9.41% for Y578S (p < 0.01). The membrane expression normalized to wt NALCN was 63.94 ± 7.26%, for L509S (p < 0.01) and 46.69 ± 7.41% for Y578S (p < 0.001). These data suggest that the observed gain-of-function effect of L509S and Y578S NALCN mutants does not result from an increased membrane expression but rather a functional enhanced activity as identified in patch-clamp experiments.
Figure 7.
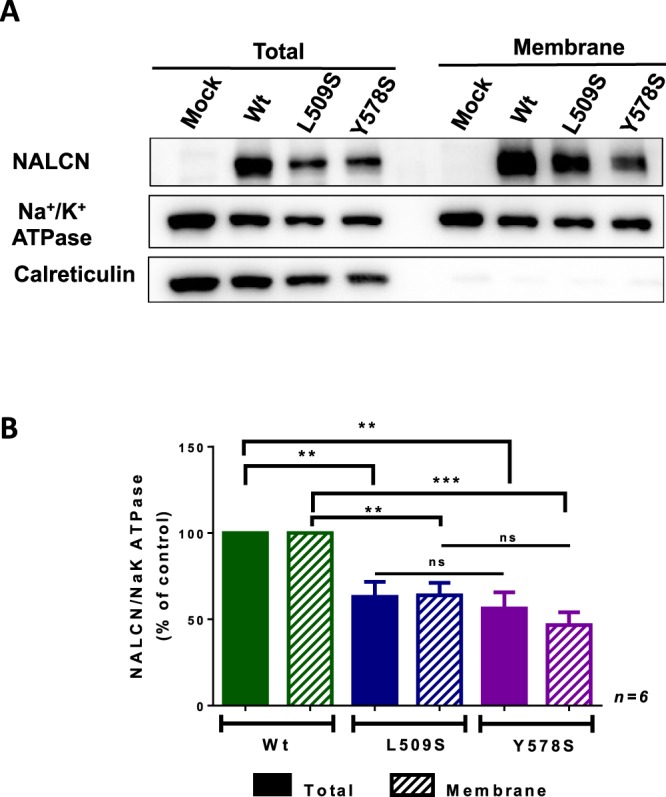
Total and membrane expression of wt NALCN channel and its L509S and Y578S variants. (A) Representative cell surface biotinylation experiment. The cell surface biotinylation assay was performed in cells transfected with NLF1 alone (Mock) or along with wt NALCN, NALCN-L509S or NALCN-Y578S (GFP tagged). The Na+/K+ ATPase, a plasma membrane protein, and the calreticulin, a reticulum-resident protein, were used internal controls. (B) The expression levels of wt NALCN, NALCN-L509S and NALCN-Y578S were normalized with the Na+/K+ ATPase level from both total and membrane fractions. The total expression compared to NALCN was: L509S, 62.99 ± 8.725%, p < 0.01; Y578S, 56.24 ± 9.408%, p < 0.01, while the membrane expression compared to NALCN was: L509S, 63.94 ± 7.261%, p < 0.01; Y578S, 46.69 ± 7.412%, p < 0.001 (n = 6 independent experiments). Statistical significance was calculated with a one-way ANOVA multiple comparisons test.
Discussion
This study describes the electrophysiological properties of recombinant NALCN channel expressed in the neuronal cell line NG108-15. In good agreement to that previously reported, the inward NALCN current was carried out by Na+ ions, blocked by Gd3+, insensitive to TTX and potentiated by lowering external Ca2+ concentration. Notably, NALCN expression in NG108-15 cells revealed novel functional properties. First, the NALCN current measured during hyperpolarizing pulses displayed inactivation-like process and a non-linear current-voltage relationship. Second, exploring representative CLIFAHDD and IHPRF1 mutations revealed that these NALCN variants showed gain- and loss of NALCN channel activity, respectively.
NALCN is mainly expressed in neurons, especially of the central nervous system (http://www.mousebrain.org/)52. It was therefore important to set-up the study of the functional properties of the wt and pathogenic variants of NALCN channel in a neuron-like environment. Functional expression of recombinant NALCN was first described using HEK293 cells3,12,32,50,51, but the relevance of this cell model to express NALCN has been disputed53,54. The pionier studies by Lu and colleagues, using HEK293 cells, have established the main NALCN electrophysiological signature, i.e. the ‘sodium leak’ properties3,12,15. The few other cellular models used to express NALCN, such as the pancreatic β MIN6 cell line13 and the undifferentiated SH-SY5Y neuronal cell line15 have confirmed some electrophysiological properties and revealed additional ones. Indeed, NALCN does not exhibit leak activity in MIN6 cells but rather an inactivating Na+ current activated by a G-protein coupled receptor (GPCR) that was evidenced following acetylcholine activation of muscarinic receptors13. In NG108-15 cells, we demonstrate that combined expression of the recombinant NALCN and NLF1 proteins was sufficient to record a Na+ background current. Importantly, the Unc79 and the Unc80 ancillary subunits are endogenously expressed in NG108-15 cells, indicating that this neuronal cell line, in which we recorded a Na+ background current, expressed the necessary components of the NALCN channelosome. Of note, the NALCN-related background Na+ current was recorded only in NALCN- and NLF1-transfected NG108-15 cells differentiated in neuron-like cells, suggesting that neuronal differentiation favors the functional expression of NALCN, possibly also by stimulating the functional expression of additional components of the neuronal NALCN channelosome.
An important finding of this study is that the Na+ background current related to heterologous NALCN expression in differentiated NG108-15 cells exhibits electrophysiological properties that have not been described elsewhere. The inward NALCN current triggered by hyperpolarizing pulse in the negative range of membrane potential (<−20 mV) displayed a time-dependent decay of the current amplitude, reminiscent of the voltage-dependent inactivation process of Na+ and Ca2+ voltage-gated channels. This inactivating current was not observed in Mock-transfected cells and was eliminated in NALCN-transfected cells by the removal of external Na+, indicating that it was linked to NALCN expression. To the best of our knowledge, such an inactivation process was never observed in the previous NALCN current recordings, especially those obtained from the HEK293 cell line that represent the vast majority of the electrophysiological data on recombinant NALCN3,12,32,50,51. This time-dependent decay of the background current is clearly attributable to the functional expression of NALCN as it was observed for both the wt and the CLIFAHDD mutants. Importantly, a significantly distinct time-course was found for the CLIFAHDD currents, which were 3 to 4 fold slower than the wt NALCN current. Also, the current-voltage (I/V) relationship of the NALCN current recorded in differentiated NG108-15 cells was not linear, contrary to that reported in published studies describing NALCN expression in HEK293 cells. These findings reveal novel electrophysiological properties of NALCN when functionally expressed in differentiated neuronal cells. It remains however important to determine if the electrophysiological properties of NALCN channel in differentiated NG108-15 cells are relevant for native neurons.
Another major finding of this study was the functional characterization of several pathogenic NALCN variants. Expression of NALCN carrying the W1287L missense mutation found in 3 IHPRF1 siblings17 did not result in any detectable Na+ background current. IHPRF1 is recessively inherited and patients carried either frameshift or missense mutations, suggesting loss-of-function mutations of NALCN. Our findings describing a loss of channel activity for the W1287L mutation of NALCN are therefore in good agreement with the genetics data. On the contrary, functional expression of the two missense mutations found in CLIFAHDD patients, L509S and Y578S, revealed a gain-of-function effect. Compared to wt NALCN, expression of the two CLIFAHDD variants showed Na+ background current of significantly higher current density. In addition, a significant slowing of the current inactivation was observed, potentially contributing also to the gain-of-function effect. The gain-of-function effect of L509S and Y578S mutations likely results from a functional alteration instead of a trafficking change since no increase, but rather a decrease, in plasma membrane expression was detected by a cell surface biotinylation assay for the two CLIFAHDD variants. Although we found no evidence for a change in Na+ selectivity or in the 2-3 fold potentiation in low external Ca2+ for the two mutants, other electrophysiological or modulation properties yet to be identified could also contribute to the gain-of-function effect of L509S and Y578S mutations.
Our data are in agreement with previous studies that suggested that pathogenic variants of NALCN linked to CLIFAHDD are gain-of-function mutations. As a matter of fact, most of the mutations found in CLIFAHDD patients localize in the transmembrane segments S5 and S6 of domains I, II, III and IV. These segments are well known to form the pore of the four-domain ion channels. Semi-dominant missense mutations found in the S6 of domain II of NCA-1 (A596V/A717V and D600E), the C. elegans ortholog of mammalian NALCN, result in an uncoordinated, and exaggerated body bends phenotype during spontaneous or stimulated locomotion (e.g. « coiler » phenotype)55. This contrasts with loss-of-function NCA-1 mutations where animals are fainters that fail to sustain sinusoidal locomotion and succumb to long periods of halting55–57. This led to the conclusion that A596V and D600E are gain-of-function mutations. In addition, synaptic calcium transients are significantly reduced in Nca loss-of-function mutants and increased in Nca gain-of-function mutants55. Another mutation was also described in the Nalcn gene in the dreamless mutant mouse50. This mutation is dominant and results in the N315L substitution in helix S6 of domain I, which is conserved among vertebrates and invertebrates. Functional expression of the N315L mutant of NALCN in HEK293 cells was found to result in a leak current significantly higher than for the wt NALCN50. Another argument that favors the gain-of-function hypothesis for the CLIFAHDD mutations comes from the fact that the EMG in a L590F patient revealed abnormalities that indicate a possible motor neuron/axon hyperexcitability27. Of note, the pan-neuronal expression of the R1230Q mutation in the NCA-1 of C. elegans that reproduces the R1181Q found in 3 patients with CLIFAHDD induced a coiling locomotion identical to that of the gain-of-function nca-1 A596V/A717V mutant22. Altogether, our data validate the hypothesis that at least some mutations found in patients with the CLIFAHDD syndrome are gain-of-function mutations.
Most of the recessive IHPRF1 (NALCN) and IHPRF2 (Unc80) mutations are predicted to result in non-functional proteins. Since we could not identify NALCN current with a NALCN variant carrying an IHPRF1 W1287L mutation in our experimental conditions, our conclusion is that this IHPRF1 missense variant of NALCN also results in a non-functional channel. It is expected that the lack of NALCN activity should lead to cellular/neuronal hyperpolarization48 and consequently a decrease in the firing properties of these cells, as observed when Nalcn is knocked out or knocked down3–10. Conversely, the two mutations found in CLIFAHDD patients, L509S and Y578S, clearly induced a gain of channel activity in the neuronal NG108-15 cells. It is predicted that higher activity of a Na+ background current should result in a more depolarized RMP, possibly in an increase of the firing properties of the NALCN-expressing cells as described in neurons from the deep mesencephalic nucleus of the dreamless mutant mouse50. However, a significant increase in NALCN activity could worsen cellular excitability by switching the RMP to depolarized state, attenuating firing activity. Strikingly, a study in which the Y578S (Y621S in C. elegans) dominant mutation was reproduced in the NCA-1 channel of C. elegans revealed a locomotor phenotype reminiscent to a loss-of-function mutation (e.g. fainting behavior). The same study reported a gain-of-function phenotype for the L509S (L556S in C. elegans) dominant mutation, e.g. coiling behavior and hypersensitivity to aldicarb27. The authors concluded that three genetic mechanisms could give rise to NALCN channelopathies: (1) NALCN loss-of function in IHPRF, (2) gain-of-function and (3) dominant-negative in dominantly inherited CLIFAHDD27. This conclusion was based on the locomotor behavior and the albicarb sensitivity of C. elegans models. Our results, at the channel/cellular level, support the two first hypothesis. First, we describe that the IHPRF mutation induces loss of NALCN channel activity. Second, we describe that both the Y578S and the L509S CLIFAHDD variants are gain-of-function mutants. In addition, we report that the Y578S variant exhibits a more pronounced gain of channel activity than the L509S variant. It is tempting to speculate that the Y578S variant would depolarize the cells to a greater extend than the L509S variant. Such a large depolarization with the Y578S mutant would result in a decrease in excitability in cell types highly depending on NALCN activity, mimicking the IHPRF loss-of-function phenotype. Such a molecular mechanism might explain why IHPRF patients and CLIFAHDD patients share several, but not all, symptoms.
To the best of our knowledge, it is the first report of functional effects of IHPRF and CLIFAHDD variants of NALCN. Although the precise mechanisms involved in the electrophysiological defects remain to be clarified, the findings reported here validate the neuronal cell line NG108-15 to investigate the functional properties of NALCN variants. Using a C. elegans model mimicking IHPRF, it was recently suggested that NALCN deficiency may be corrected by pharmacological targeting of other channels58. Conversely, one may predict that partial inhibition of NALCN could present some benefits for CLIFAHDD patients. Cellular systems expressing IHPRF and CLIFAHDD variants are therefore of interest for further pharmacological investigations. As a conclusion, our present work provides the first report of functional impact of pathogenic variants of NALCN found in the two-associated and devastating human diseases, and paves the way to the identification of therapeutical strategies to treat NALCN-related diseases.
Acknowledgements
The authors thank Dr. Mei Zhen and Dr. Kayota Aoyagi for the kind gift of the NLF1-encoding cDNA, as well as Drs Isabelle Bidaud, Sophie Nicole, Jean Chemin, Nathalie Guérineau and Adriano Senatore for technical support and critical reading of the manuscript. This work was supported by Labex ‘Ion Channel Science and Therapeutics’, Fondation Maladies Rares (FMR), Association Française contre les Myopathies (AFM) and Fondation pour la Recherche Médicale (FRM). Hathaichanok Impheng is co-funded by both Naresuan University (Thailand) and the French Embassy in Thailand.
Author Contributions
M.B., H.I. and Z.S. conducted the experiments. M.B., H.I., Z.S., P.L. and A.M. analyzed the data. P.L. and A.M. conceptualized and designed experiments. P.L. and A.M. prepared the manuscript. Project administration and funding acquisition were done by P.L. and A.M. All authors read and approved the final manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Malik Bouasse and Hathaichanok Impheng contributed equally.
References
- 1.Hille, B. Ion channels of excitable membranes, 3rd ed. Sinauer Associates, Sunderland, MA, (2001).
- 2.Kandel, E. R., Schwartz, J. H., Jessel, T. M., Siegelbaum, S. A. & Hudspeth, A. J. Principles of Neural Science. 4th ed. New York, NY: McGraw Hill (2013).
- 3.Lu B, et al. The neuronal channel NALCN contributes resting sodium permeability and is required for normal respiratory rhythm. Cell. 2007;129:371–383. doi: 10.1016/j.cell.2007.02.041. [DOI] [PubMed] [Google Scholar]
- 4.Lu TZ, Feng ZP. A sodium leak current regulates pacemaker activity of adult central pattern generator neurons in Lymnaea stagnalis. PLoS One. 2011;6:e18745. doi: 10.1371/journal.pone.0018745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xie L, et al. NLF-1 delivers a sodium leak channel to regulate neuronal excitability and modulate rhythmic locomotion. Neuron. 2013;77:1069–1082. doi: 10.1016/j.neuron.2013.01.018. [DOI] [PubMed] [Google Scholar]
- 6.Gao S, et al. The NCA sodium leak channel is required for persistent motor circuit activity that sustains locomotion. Nat Commun. 2015;6:6323. doi: 10.1038/ncomms7323. [DOI] [PubMed] [Google Scholar]
- 7.Flourakis M, et al. A Conserved Bicycle Model for Circadian Clock Control of Membrane Excitability. Cell. 2015;162:836–848. doi: 10.1016/j.cell.2015.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lutas, A., Lahmann, C., Soumillon, M. & Yellen, G. The leak channel NALCN controls tonic firing and glycolytic sensitivity of substantia nigra pars reticulata neurons. Elife5, 10.7554/eLife.15271 (2016). [DOI] [PMC free article] [PubMed]
- 9.Shi Y, et al. Nalcn Is a “Leak” Sodium Channel That Regulates Excitability of Brainstem Chemosensory Neurons and Breathing. J Neurosci. 2016;36:8174–8187. doi: 10.1523/JNEUROSCI.1096-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yeh SY, et al. Respiratory Network Stability and Modulatory Response to Substance P Require Nalcn. Neuron. 2017;94:294–303 e294. doi: 10.1016/j.neuron.2017.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ford, N. C., Ren, D. & Baccei, M. L. NALCN channels enhance the intrinsic excitability of spinal projection neurons. Pain, 10.1097/j.pain.0000000000001258 (2018). [DOI] [PMC free article] [PubMed]
- 12.Lu B, et al. Peptide neurotransmitters activate a cation channel complex of NALCN and UNC-80. Nature. 2009;457:741–744. doi: 10.1038/nature07579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swayne LA, et al. The NALCN ion channel is activated by M3 muscarinic receptors in a pancreatic beta-cell line. EMBO Rep. 2009;10:873–880. doi: 10.1038/embor.2009.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim BJ, et al. Involvement of Na(+)-leak channel in substance P-induced depolarization of pacemaking activity in interstitial cells of Cajal. Cell Physiol Biochem. 2012;29:501–510. doi: 10.1159/000338504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu B, et al. Extracellular calcium controls background current and neuronal excitability via an UNC79-UNC80-NALCN cation channel complex. Neuron. 2010;68:488–499. doi: 10.1016/j.neuron.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Philippart, F. & Khaliq, Z. M. Gi/o protein-coupled receptors in dopamine neurons inhibit the sodium leak channel NALCN. Elife7, 10.7554/eLife.40984 (2018). [DOI] [PMC free article] [PubMed]
- 17.Al-Sayed MD, et al. Mutations in NALCN cause an autosomal-recessive syndrome with severe hypotonia, speech impairment, and cognitive delay. Am J Hum Genet. 2013;93:721–726. doi: 10.1016/j.ajhg.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koroglu C, Seven M, Tolun A. Recessive truncating NALCN mutation in infantile neuroaxonal dystrophy with facial dysmorphism. J Med Genet. 2013;50:515–520. doi: 10.1136/jmedgenet-2013-101634. [DOI] [PubMed] [Google Scholar]
- 19.Gal M, et al. A novel homozygous splice site mutation in NALCN identified in siblings with cachexia, strabismus, severe intellectual disability, epilepsy and abnormal respiratory rhythm. Eur J Med Genet. 2016;59:204–209. doi: 10.1016/j.ejmg.2016.02.007. [DOI] [PubMed] [Google Scholar]
- 20.Takenouchi T, et al. Biallelic mutations in NALCN: Expanding the genotypic and phenotypic spectra of IHPRF1. Am J Med Genet A. 2018;176:431–437. doi: 10.1002/ajmg.a.38543. [DOI] [PubMed] [Google Scholar]
- 21.Chong JX, et al. De novo mutations in NALCN cause a syndrome characterized by congenital contractures of the limbs and face, hypotonia, and developmental delay. Am J Hum Genet. 2015;96:462–473. doi: 10.1016/j.ajhg.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aoyagi K, et al. A Gain-of-Function Mutation in NALCN in a Child with Intellectual Disability, Ataxia, and Arthrogryposis. Hum Mutat. 2015;36:753–757. doi: 10.1002/humu.22797. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, et al. A de novo mutation in the NALCN gene in an adult patient with cerebellar ataxia associated with intellectual disability and arthrogryposis. Clin Genet. 2016;90:556–557. doi: 10.1111/cge.12851. [DOI] [PubMed] [Google Scholar]
- 24.Fukai R, et al. De novo missense mutations in NALCN cause developmental and intellectual impairment with hypotonia. J Hum Genet. 2016;61:451–455. doi: 10.1038/jhg.2015.163. [DOI] [PubMed] [Google Scholar]
- 25.Karakaya M, et al. Novel Mutations in the Nonselective Sodium Leak Channel (NALCN) Lead to Distal Arthrogryposis with Increased Muscle Tone. Neuropediatrics. 2016;47:273–277. doi: 10.1055/s-0036-1584084. [DOI] [PubMed] [Google Scholar]
- 26.Sivaraman I, Friedman NR, Prayson RA. Muscle biopsy findings in a child with NALCN gene mutation. J Clin Neurosci. 2016;34:222–223. doi: 10.1016/j.jocn.2016.06.018. [DOI] [PubMed] [Google Scholar]
- 27.Bend EG, et al. NALCN channelopathies: Distinguishing gain-of-function and loss-of-function mutations. Neurology. 2016;87:1131–1139. doi: 10.1212/WNL.0000000000003095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lozic B, et al. Novel NALCN variant: altered respiratory and circadian rhythm, anesthetic sensitivity. Ann Clin Transl Neurol. 2016;3:876–883. doi: 10.1002/acn3.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vivero M, et al. Additional de novo missense genetic variants in NALCN associated with CLIFAHDD syndrome. Clin Genet. 2017;91:929–931. doi: 10.1111/cge.12899. [DOI] [PubMed] [Google Scholar]
- 30.Perez Y, et al. UNC80 mutation causes a syndrome of hypotonia, severe intellectual disability, dyskinesia and dysmorphism, similar to that caused by mutations in its interacting cation channel NALCN. J Med Genet. 2016;53:397–402. doi: 10.1136/jmedgenet-2015-103352. [DOI] [PubMed] [Google Scholar]
- 31.Shamseldin HE, et al. Mutations in UNC80, Encoding Part of the UNC79-UNC80-NALCN Channel Complex, Cause Autosomal-Recessive Severe Infantile Encephalopathy. Am J Hum Genet. 2016;98:210–215. doi: 10.1016/j.ajhg.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stray-Pedersen A, et al. Biallelic Mutations in UNC80 Cause Persistent Hypotonia, Encephalopathy, Growth Retardation, and Severe Intellectual Disability. Am J Hum Genet. 2016;98:202–209. doi: 10.1016/j.ajhg.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valkanas E, et al. Phenotypic evolution of UNC80 loss of function. Am J Med Genet A. 2016;170:3106–3114. doi: 10.1002/ajmg.a.37929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chérot E., Keren B., Dubourg C., Carré W., Fradin M., Lavillaureix A., Afenjar A., Burglen L., Whalen S., Charles P., Marey I., Heide S., Jacquette A., Heron D., Doummar D., Rodriguez D., Billette de Villemeur T., Moutard M.-L., Guët A., Xavier J., Périsse D., Cohen D., Demurger F., Quélin C., Depienne C., Odent S., Nava C., David V., Pasquier L., Mignot C. Using medical exome sequencing to identify the causes of neurodevelopmental disorders: Experience of 2 clinical units and 216 patients. Clinical Genetics. 2017;93(3):567–576. doi: 10.1111/cge.13102. [DOI] [PubMed] [Google Scholar]
- 35.Angius A., Cossu S., Uva P., Oppo M., Onano S., Persico I., Fotia G., Atzeni R., Cuccuru G., Asunis M., Cucca F., Pruna D., Crisponi L. Novel NALCN biallelic truncating mutations in siblings with IHPRF1 syndrome. Clinical Genetics. 2018;93(6):1245–1247. doi: 10.1111/cge.13162. [DOI] [PubMed] [Google Scholar]
- 36.Obeid Tasneem, Hamzeh Abdul Rezzak, Saif Fatima, Nair Pratibha, Mohamed Madiha, Al-Ali Mahmoud Taleb, Bastaki Fatma. Identification of a novel homozygous UNC80 variant in a child with infantile hypotonia with psychomotor retardation and characteristic facies-2 (IHPRF2) Metabolic Brain Disease. 2018;33(3):869–873. doi: 10.1007/s11011-018-0200-z. [DOI] [PubMed] [Google Scholar]
- 37.Campbell J, et al. NALCN Dysfunction as a Cause of Disordered Respiratory Rhythm With Central Apnea. Pediatrics. 2018;141:S485–S490. doi: 10.1542/peds.2017-0026. [DOI] [PubMed] [Google Scholar]
- 38.Bourque Danielle K., Dyment David A., MacLusky Ian, Kernohan Kristin D., McMillan Hugh J. Periodic breathing in patients with NALCN mutations. Journal of Human Genetics. 2018;63(10):1093–1096. doi: 10.1038/s10038-018-0484-1. [DOI] [PubMed] [Google Scholar]
- 39.Hong H, et al. Case Report of Pediatric Channelopathies With UNC80 and KCNJ11 Mutations Having Abnormal Respiratory Control Treated With Positive Airway Pressure Therapy. J Clin Sleep Med. 2018;14:1419–1425. doi: 10.5664/jcsm.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bramswig Nuria C., Bertoli-Avella Aida M., Albrecht Beate, Al Aqeel Aida I., Alhashem Amal, Al-Sannaa Nouriya, Bah Maissa, Bröhl Katharina, Depienne Christel, Dorison Nathalie, Doummar Diane, Ehmke Nadja, Elbendary Hasnaa M., Gorokhova Svetlana, Héron Delphine, Horn Denise, James Kiely, Keren Boris, Kuechler Alma, Ismail Samira, Issa Mahmoud Y., Marey Isabelle, Mayer Michèle, McEvoy-Venneri Jennifer, Megarbane Andre, Mignot Cyril, Mohamed Sarar, Nava Caroline, Philip Nicole, Ravix Cecile, Rolfs Arndt, Sadek Abdelrahim Abdrabou, Segebrecht Lara, Stanley Valentina, Trautman Camille, Valence Stephanie, Villard Laurent, Wieland Thomas, Engels Hartmut, Strom Tim M., Zaki Maha S., Gleeson Joseph G., Lüdecke Hermann-Josef, Bauer Peter, Wieczorek Dagmar. Genetic variants in components of the NALCN–UNC80–UNC79 ion channel complex cause a broad clinical phenotype (NALCN channelopathies) Human Genetics. 2018;137(9):753–768. doi: 10.1007/s00439-018-1929-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Angius A, et al. Exome sequencing in Crisponi/cold-induced sweating syndrome-like individuals reveals unpredicted alternative diagnoses. Clin Genet. 2019;95:607–614. doi: 10.1111/cge.13532. [DOI] [PubMed] [Google Scholar]
- 42.Carneiro TN, et al. Utility of trio-based exome sequencing in the elucidation of the genetic basis of isolated syndromic intellectual disability: illustrative cases. Appl Clin Genet. 2018;11:93–98. doi: 10.2147/TACG.S165799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuptanon C, et al. Whole exome sequencing revealed mutations in FBXL4, UNC80, and ADK in Thai patients with severe intellectual disabilities. Gene. 2019;696:21–27. doi: 10.1016/j.gene.2019.01.049. [DOI] [PubMed] [Google Scholar]
- 44.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chemin, J., Nargeot, J. & Lory, P. Neuronal T-type alpha 1H calcium channels induce neuritogenesis and expression of high-voltage-activated calcium channels in the NG108-15 cell line. J Neurosci22, 6856–6862, doi:20026671 (2002). [DOI] [PMC free article] [PubMed]
- 46.Monteil A, et al. Inhibition of Cav3.2 T-type Calcium Channels by Its Intracellular I-II Loop. J Biol Chem. 2015;290:16168–16176. doi: 10.1074/jbc.M114.634261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duhr F, et al. Cdk5 induces constitutive activation of 5-HT6 receptors to promote neurite growth. Nat Chem Biol. 2014;10:590–597. doi: 10.1038/nchembio.1547. [DOI] [PubMed] [Google Scholar]
- 48.Cochet-Bissuel M, Lory P, Monteil A. The sodium leak channel, NALCN, in health and disease. Front Cell Neurosci. 2014;8:132. doi: 10.3389/fncel.2014.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reinl EL, Cabeza R, Gregory IA, Cahill AG, England SK. Sodium leak channel, non-selective contributes to the leak current in human myometrial smooth muscle cells from pregnant women. Mol Hum Reprod. 2015;21:816–824. doi: 10.1093/molehr/gav038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Funato H, et al. Forward-genetics analysis of sleep in randomly mutagenized mice. Nature. 2016;539:378–383. doi: 10.1038/nature20142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eigenbrod O, et al. Rapid molecular evolution of pain insensitivity in multiple African rodents. Science. 2019;364:852–859. doi: 10.1126/science.aau0236. [DOI] [PubMed] [Google Scholar]
- 52.Zeisel A, et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- 53.Boone AN, Senatore A, Chemin J, Monteil A, Spafford JD. Gd3+ and calcium sensitive, sodium leak currents are features of weak membrane-glass seals in patch clamp recordings. PLoS One. 2014;9:e98808. doi: 10.1371/journal.pone.0098808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Egan JM, Peterson CA, Fry WM. Lack of current observed in HEK293 cells expressing NALCN channels. Biochim Open. 2018;6:24–28. doi: 10.1016/j.biopen.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yeh E, et al. A putative cation channel, NCA-1, and a novel protein, UNC-80, transmit neuronal activity in C. elegans. PLoS Biol. 2008;6:e55. doi: 10.1371/journal.pbio.0060055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sedensky MM, Meneely PM. Genetic analysis of halothane sensitivity in Caenorhabditis elegans. Science. 1987;236:952–954. doi: 10.1126/science.3576211. [DOI] [PubMed] [Google Scholar]
- 57.Humphrey JA, et al. A putative cation channel and its novel regulator: cross-species conservation of effects on general anesthesia. Curr Biol. 2007;17:624–629. doi: 10.1016/j.cub.2007.02.037. [DOI] [PubMed] [Google Scholar]
- 58.Kasap M, Bonnett K, Aamodt EJ, Dwyer DS. Akinesia and freezing caused by Na(+) leak-current channel (NALCN) deficiency corrected by pharmacological inhibition of K(+) channels and gap junctions. J Comp Neurol. 2017;525:1109–1121. doi: 10.1002/cne.24119. [DOI] [PubMed] [Google Scholar]