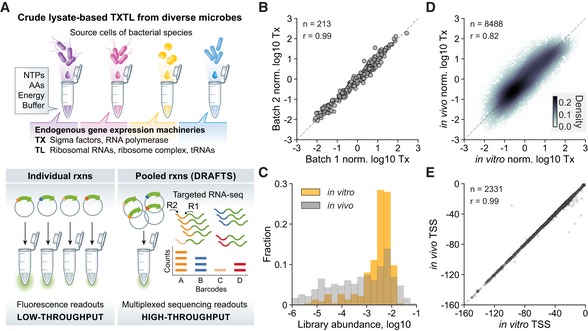
DRAFTS uses crude cell lysate‐based cell‐free expression systems to harness source host cell's endogenous gene expression machineries. Compared to conventional single‐channel reporters with color or fluorescence readouts, multiplexed sequencing readouts from pooled reactions in DRAFTS scale up the throughput of measurement.
Biological replicates of transcriptional profiles (Tx) measured in separately prepared cell‐free lysate batches of E. coli.
Comparison of abundances of each library constructs in in vivo and in vitro measurements.
Correlation between transcriptional profiles (Tx) for regulatory sequence libraries from in vitro and in vivo measurements in E. coli. The color scale indicates the density of data points in a given area of the plot.
Correlation between primary TSS calls (in bp from ATG start codon) of regulatory sequences from in vitro and in vivo measurements in E. coli.
Data information: Dashed lines represent
y =
x in (B), (D), and (E). Sample sizes (
n) and Pearson correlation coefficients (
r) are shown in each plot. For normalization, transcription levels in log
10 scale were transformed to
Z‐scores. All measurements except (C) are based on two biological replicates.
Source data are available online for this figure.