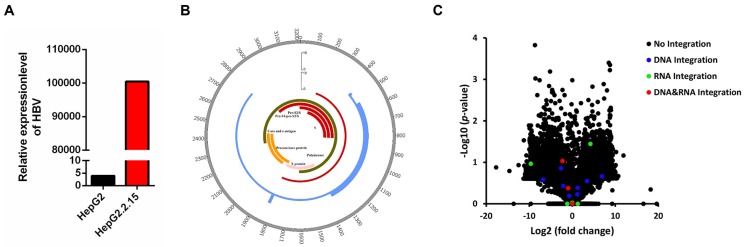
Figure 1.
HBV integration breakpoints distribution in HBV genome and gene expression change in HepG2.2.15. (A) Detection of HBV gene copy number by PCR. (B) Distribution of integration breakpoints in the HBV genome in HepG2.2.15 (genome integration breakpoints of HepG2.2.15: blue, transcriptome integration breakpoints of HepG2.2.15: red) are shown. Histograms were constructed for 100bp intervals. HBV genes with different functions are shown. (C) Volcano plot of differently expressed genes between HepG2.2.15 and HepG2. The logarithms of the fold changes of genes (x-axis) are plotted against the negative logarithm of their p-value(y-axis). Black dots represent genes without HBV integration (No integration), blue dots represent genes with HBV integration in genome (DNA integration), green dots represent genes with HBV integration in transcriptome (RNA integration), red dots represent genes with HBV integration in both genome and transcriptome (DNA&RNA integration).