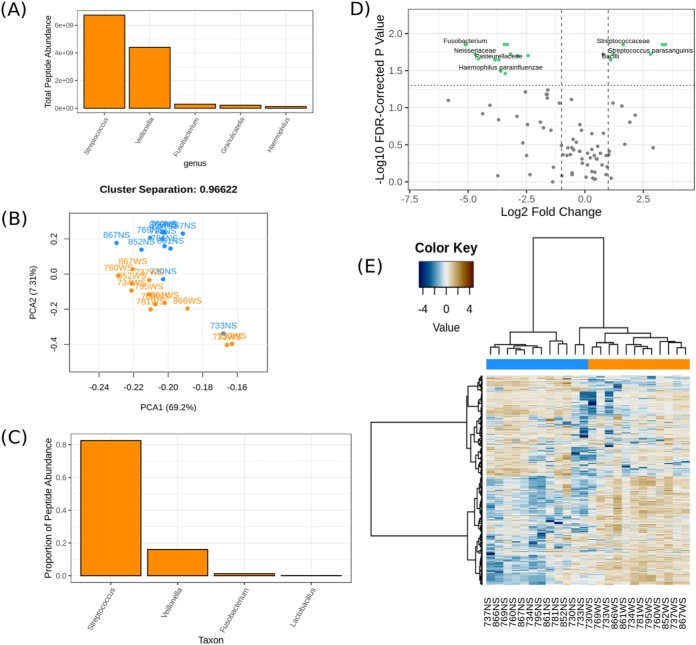
Fig. 4.
A sampling of metaQuantome visualizations for the oral microbiome dataset. (A) The five most abundant genera in the WS (sugar-pulsed) condition. (B) A principal component analysis on functional abundance separates NS (blue) and WS samples (orange), with some outliers. The separation between the clusters can be seen in the title and is defined in Equation 1. (C) Proportion of total peptide abundance in WS attributed to genera contributing to carbohydrate metabolism (GO:0005975). (D) A volcano plot representing the results of the taxonomic differential abundance analysis, with the fold change reported as abundance in WS over abundance in NS. Taxa with a statistically significant fold change at a user-defined alpha (here, 0.05) are shown with green dots and labeled (some labels removed to reduce overplotting). (E) A hierarchically clustered heatmap of functional annotations separates NS (blue) and WS (orange) samples.