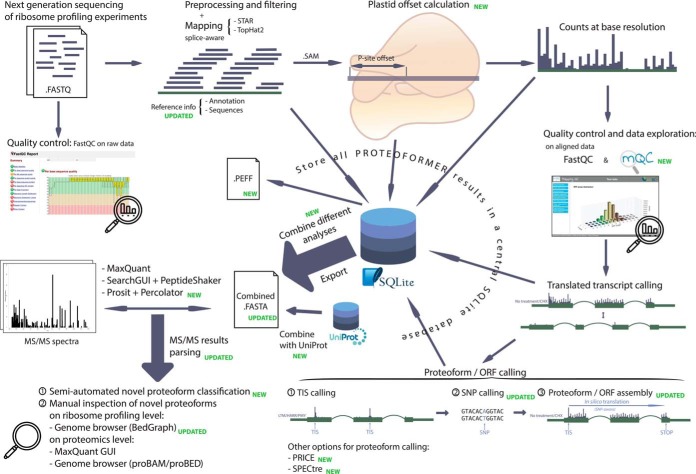
Fig. 1.
Most important parts of the PROTEOFORMER pipeline workflow. The pipeline starts with raw reads from a ribosome profiling experiment, provided in FASTQ format. The quality of these raw reads are checked with FastQC (40). Next, the reads are preprocessed, filtered and mapped to the reference genome. By using P-site offsets (calculated with Plastid (28)), these alignments can be pinpointed at the base level. Quality of the alignments and general data outlook will be checked with help of FastQC (40) and mQC (29). If the user is satisfied with the output, one can continue with the pipeline. PROTEOFORMER will search for the transcript isoforms with translation evidence. Based on these, the translated proteoforms can be deduced. The workflow used in the previous PROTEOFORMER (11) version can be applied (TIS calling, SNP calling and proteoform assembly) or one can use PRICE (12) or SPECtre (13) to determine these proteoforms. All results of these earlier steps are saved in an SQLite results database. For MS-based validation, the results can be exported, combined and even merged with canonical information from UniProt. The end result is a FASTA file that can be used for database searching of MS/MS spectra with tools like MaxQuant (19), SearchGUI-PeptideShaker (63, 64) or Prosit (24) in combination with Percolator (25). Several novel scripts were added to the pipeline to use these search results for counting database hits and classifying new proteoforms and novel translation events in a semi-automated fashion. Identifications can also be manually inspected on both ribosome (e.g. by browsing the PROTEOFORMER BedGraph files in a genome browser environment) and MS level (MS software interface or converted MS identification files in proBAM/proBED format (57, 58) in the same genome browser session as the BedGraph ribofiles).