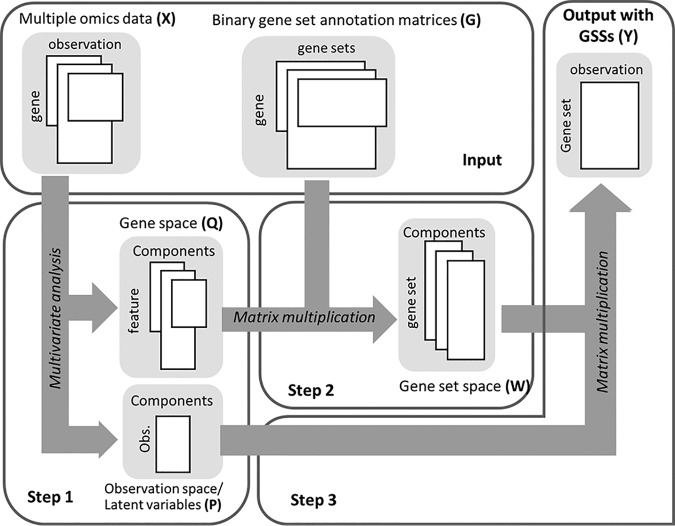
Fig. 1.
Schematic representation of the MOGSA algorithm. The algorithm requires pairs of matrices as input; multiple omics data matrices and corresponding gene-set (GS) annotation matrices. In step 1, the multiple matrices are analyzed using a multivariate analysis (MVA) method resulting in an observation space and gene space. Next, the gene-set annotation matrices are projected on the same space, and the resulting matrix contains the gene-set space. The last step is to reconstruct gene-set-observation by multiplying the observation and gene-set spaces.