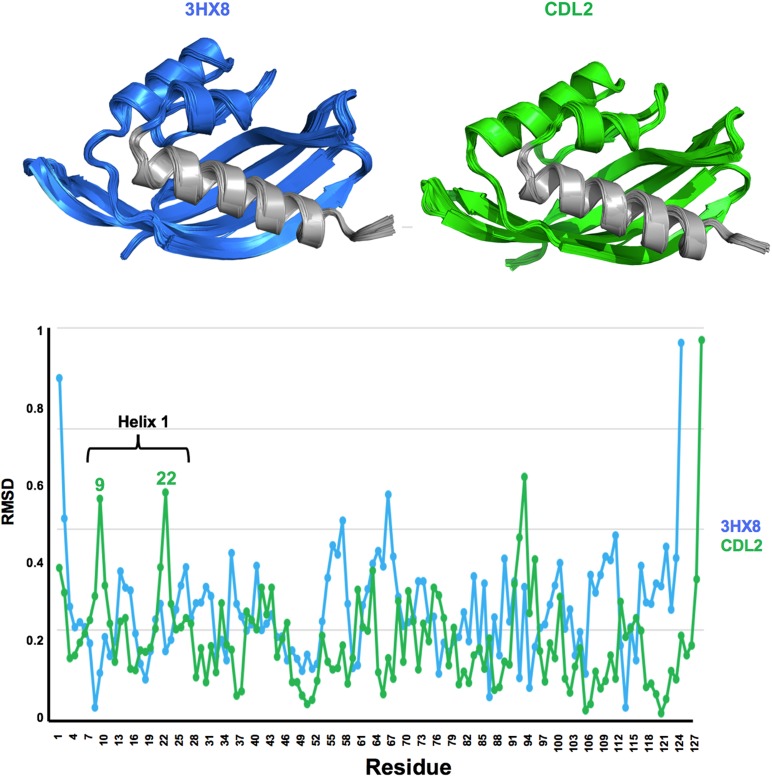
Fig. 8.
Molecular dynamics simulations of PDB 3HX8 and design CDL2. A small shearing movement of the N-terminal helix, in the same direction as that observed in our crystal structures (Fig. 8) is observed for the CDL2 starting model, with peaks in RMSD backbone shifts corresponding to residues near the two ends of the helix.