Abstract
Pancreatic ductal adenocarcinoma (PDAC) adopts several unique metabolic strategies to support primary tumor growth. Whether additional metabolic strategies are adopted to support metastatic tumorigenesis is less clear. This could be particularly relevant for distant metastasis, which often follows a rapidly progressive clinical course. Here we report that PDAC distant metastases evolve a unique series of metabolic reactions to maintain activation of the anabolic glucose enzyme phosphogluconate dehydrogenase (PGD). PGD catalytic activity was recurrently elevated across distant metastases, and modulating PGD activity levels dictated tumorigenic capacity. Metabolomics data raised the possibility that distant metastases evolved a core pentose conversion pathway (PCP) that converted glucose-derived metabolites into PGD substrate, thereby hyperactivating the enzyme. Consistent with this, each individual metabolite in the PCP stimulated PGD catalysis in distant metastases, and knockdown of each individual PCP enzyme selectively impaired tumorigenesis. We propose that the PCP manufactures PGD substrate outside of the rate-limiting oxidative pentose phosphate pathway (oxPPP). This enables PGD-dependent tumorigenesis by providing adequate substrate to fuel high catalytic activity, and raises the possibility that PDAC distant metastases adopt their own unique metabolic strategies to support tumor growth.
Introduction
Pancreatic ductal adenocarcinoma (PDAC) remains one of the most lethal of all human malignancies, and is projected to become the second leading cause of cancer deaths in the coming years [1]. That is because most patients develop widely metastatic disease that progresses rapidly and is treatment-resistant [2, 3].
PDAC evolves through well-defined stages of progression: benign in situ precursor lesions (for example, PanINs), invasive primary tumor growth, subclonal evolution, metastatic spread, and patient death [4, 5]. Approximately 70% of PDAC patients die of widely metastatic disease [6, 7]. One form of metastatic spread is intra-abdominal deposits (peritoneal carcinomatosis). The other is distant metastasis [7]. Distant metastasis is a terminal process whereby a subset of malignant cells exit the primary tumor, disseminate in the bloodstream, colonize other organs, and successfully form treatment-resistant metastatic tumors there [8]. In PDAC patients, the distant metastatic disease often presents suddenly and progresses rapidly: most patients are either initially diagnosed with widespread distant metastasis or develop it soon thereafter, and death ensues within weeks to months [3, 9]. This also extends to patients without evidence of metastasis at diagnosis: over half are dead of widespread distant metastasis within 12–24 months [7, 9].
The accelerated progression of PDAC distant metastasis remains enigmatic. Although tumor cell dissemination begins early in progression [10], clinically-relevant distant metastases appear late and are seeded by the latest-appearing subclones that evolve within the primary pancreatic tumor [11]. Once established, distant metastases progress rapidly despite relatively modest proliferation rates [11–13]. At autopsy, this manifests as hundreds to thousands of grossly visible metastatic tumors that diffusely involve the liver and/or lungs [6, 7]. Although exhaustive efforts have attempted to identify metastasis-specific genetic alterations that might drive this process, all known driver mutations [11, 12, 14–17] and consequential copy number changes [15–20] are shared between the primary tumor and metastases, including those required for metastasis itself [21–24]. Thus, it remains unclear whether additional mechanisms are selected during disease evolution to uniquely support metastatic tumorigenesis.
We recently reported that human PDACs evolved strong phosphogluconate dehydrogenase (PGD) dependence during the evolution of distant metastasis [13]. PGD is a biosynthetic enzyme that converts glucose-derived substrates into NADPH and pentose riboside precursors. PGD loss-of-function selectively reversed anabolic glucose metabolism, reprogrammed epigenetic state, malignant gene expression, and tumorigenesis in PDAC distant metastatic subclones with no effect on PGD-independent controls [13]. It is well-established that primary PDACs rely on several unusual metabolic adaptations to support tumorigenesis within the dense and nutrient-poor primary tumor stroma [25–37], which may itself act as a barrier against metastasis [32]. Here we report that human PDAC distant metastases rely on an equally unusual series of pentose conversion reactions to support PGD-dependent tumorigenesis.
Results and discussion
We previously identified PGD dependence in PDAC distant metastases using a unique panel of clonal cell lines isolated from PDAC patients by rapid autopsies. These low passage cells are particularly suited to investigate human PDAC evolution, since they represent sequence-verified subclones that faithfully retain the morphologic, genetic, epigenomic, transcriptomic, and phenotypic characteristics of the parental tissues from which they were derived [6, 7, 12, 13, 15]. As such, these samples were employed to further investigate the mechanisms whereby PGD dependence was selected during the evolution of distant metastasis in these patients.
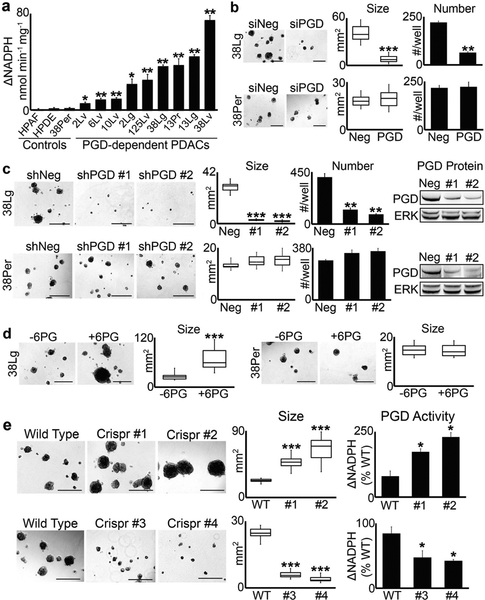
We initially identified PGD as a potentially important pro-metastatic driver gene because steady state levels of PGD substrate (6-phosphogluconate: 6PG) were strongly and recurrently depleted across distant metastatic subclones by liquid chromatography followed by high resolution mass spectrometry (LC–HRMS) metabolomics experiments [13]. Because PGD is not genetically altered [11, 12, 15, 17] and expression levels did not explain these findings [12, 13], we hypothesized that substrate depletion reflected high catalytic activity [13]. To more formally test this hypothesis, we measured PGD catalytic activity within cell extracts harvested from PGD-dependent distant metastases [13] and compared activity levels to PGD-independent control cells [13]. These experiments revealed that PGD-dependent subclones possessed variable yet recurrently elevated endogenous catalytic activity relative to control cells (Fig. 1a). This included matched liver and lung metastatic subclones that had diverged from a peritoneal subclone in the same patient (patient 38), a primary tumor subclone that seeded a matched lung metastasis in another patient (patient 13), matched liver and lung metastases from yet another patient (patient 2), and individual distant metastases isolated from additional patients. Thus, PGD catalytic activity was recurrently elevated in distant metastatic subclones isolated from different PDAC patients and in phylogenetically-related subclones that evolved within the same patient(s).
Fig. 1.
PGD enzyme activity dictates tumorigenic capacity. a PGD enzyme assays were performed on cell extracts isolated from each low passage (<15), sequence-verified clonal PDAC distant metastasis, and control cells as indicated. For all 2D culture expansions, cells were grown in DMEM with 10% FBS and were free of mycoplasma (Sigma LookOut). For enzyme assays, cells were lysed and the extracts filtered through Amicon 10K columns to remove endogenous metabolites. Protein concentration was then measured with BCA (Pierce), and 10 μg of protein (in 10 μl) was incubated with 0.2 mM NADP (Sigma) and 0.4 mM 6PG (Sigma) in 90 μl of reaction buffer (50 mM Tris, pH = 8; 1 mM MgCl2; complete EDTA-free protease inhibitors) in a 96-well plate. NADPH absorbance (340 nm) was measured over 10 min and nmol NADPH produced was determined from a standard curve. For these and all subsequent enzyme assay experiments, measurements were performed in triplicate, nonspecific background absorbance was subtracted from each replicate, and NADPH production rates (ΔNADPH) were calculated as nmol NADPH/min/mg protein. b 38Lg and 38Per cells were transfected with control (siNeg) or PGD siRNAs as described [13], trypsinized into single cells, and grown in 3D suspension Matrigel assays, also as described in [13]. For these and all other Matrigel suspension assays, 4000 cells/well were plated into low attachment 24-well plates in quadruplicate, and tumoroid growth monitored over a period of approximately 3 weeks with 100 μl media (DMEM with 5% Matrigel and 2% FBS) feeds every 5–7 days. Tumor numbers were counted at the end of the experiment, and tumor area was quantified using CellSens software (n = 15 tumors across 5–10 microscopic fields). c Stable 38Lg and 38Per cell lines were generated by lentiviral transduction and puromycin selection using non-targeting control shRNAs (Sigma) and two separate PGD shRNAs (shRNA #1: genic region; shRNA #2: 3′-UTR; Sigma TRCN274974 and TRCN274976). Although proliferation rates in 2D were not impaired (data not shown), 3D tumoroid growth was significantly impaired in 38Lg shPGD cells. Western blots demonstrated efficient knockdown of PGD protein in 38Lg and the 38Per controls, with no effects on total ERK 1/2 levels (antibodies: Cell Signaling 13389 and 4696). d 38Lg and 38Per cells were plated in 3D with complete media supplemented with (+6PG) or without (−6PG) 5 mM 6PG. 6PG significantly increased tumoroid size in 38Lg. e Four separate Crispr clones (#1, #2, #3, and #4) were generated for 38Lg with viral transduction followed by puromycin selection using a Crisper sgRNA targeted to exon 8, which encodes the 6PG substrate binding region (Sigma LentiCrispr, ccctggaatacggcgtacccgtc). Effects on 3D tumoroid size mirrored PGD catalytic activity, as measured by PGD enzyme assays. For all experiments in this figure, statistical significance was calculated using 2-tailed t-tests (*p < 0.05; **p ≤ 0.001; ***p ≤ 0.0001)
Our previous studies [13] showed that PGD knockdown with siRNAs or PGD inhibition with 6-aminonicotinamide (6AN) [38] recurrently impaired the intrinsic ability of distant metastatic subclones to form homotypic tumors (tumoroids) in 3D experimental platforms, with no effect on PGD-independent controls. 3D tumoroid assays [13, 39–41] were required to investigate the tumorigenic properties of rapid autopsy subclones, since these cells did not effectively form metastatic tumors in immunodeficient mice [13]. We therefore wished to extend these findings by more fully examining the consequences of modulating PGD function on tumorigenic capacity. First, shRNA approaches were used to generate stable PGD knockdown in a lung metastasis (38Lg: PGD-dependent) and a matched peritoneal deposit (38Per: PGD-independent) isolated from the same patient. Similar to our previous observations by acute knockdown with siRNA [13] (for example, Fig. 1b), stable shRNA knockdown strongly impaired 3D tumoroid growth in 38Lg but not 38Per (Fig. 1c). To determine if RNAi effects could be rescued by physiologically relevant re-expression of PGD, full-length exogenous PGD was introduced and expressed to modest levels in 38Lg (Supplementary Fig. 1a), followed by knockdown of the endogenous gene with siRNAs targeted to the 3′-UTR. Because exogenous PGD transcripts were modestly expressed and resistant to 3′-UTR knockdown, exogenous PGD expression was retained at physiologic levels that were higher than knockdown of the endogenous gene yet still comparable with or even lower than baseline controls during RNAi experiments (Supplementary Fig. 1b). Consistent with a physiologic rescue, exogenous PGD fully or near-fully restored 3D tumoroid growth in response to multiple 3′-UTR siRNAs (Supplementary Fig. 1c). Having established that RNAi loss-of-function strongly impaired tumorigenesis, we next tested if provision of exogenous 6PG substrate might enhance tumorigenesis, since 6PG is rate-limiting for high PGD activity. Indeed, 6PG greatly increased the size of 38Lg tumoroids with no effect on 38Per (Fig. 1d). Finally, we generated stable PGD mutant lines for 38Lg using Crispr/Cas small guide RNA (sgRNA) targeted to the PGD substrate binding region. This strategy was employed because Crispr mutations in specific domains more precisely modulate gene function than conventional 5′-truncations [42]. Remarkably, the same sgRNA sequence generated a mixture of clones that possessed gain-of-function or loss-of-function tumorigenic phenotypes in 3D that closely mirrored their effects on PGD catalytic activity (Fig. 1e) and protein expression (Supplementary Fig. 1d). Sequencing of PGD mRNA transcripts from these clones detected heterozygous frameshift rearrangements in loss-of-function clones and similar point mutations in gain-of-function clones that were all present within the region targeted by the sgRNA (Supplementary Fig. 1e). Collectively, these findings validated our previous results [13], and strongly suggested that PGD functional status is a key determinant of tumorigenic capacity.
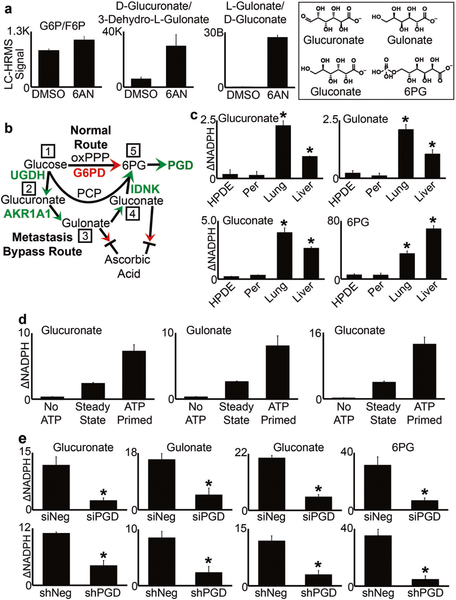
There are several diverse biochemical mechanisms that may act directly on the PGD protein itself to regulate catalytic activity [43–49], which could vary among different patients [13]. However, none are functional without provision of adequate substrate, which is rate-limiting as reflected in our metabolomics data [13] and by 6PG supplementation in 3D (Fig. 1d). The oxidative pentose phosphate pathway (oxPPP) is the canonical route to 6PG production in normal and neoplastic cells, but is itself rate-limited by an upstream G6PD gatekeeper step that is subject to tight negative feedback controls [50]. Thus, G6PD restricts how much 6PG can be produced to fuel PGD catalysis through the oxPPP. Furthermore, G6PD is dispensable in mouse models of PDAC [37], and LC–HRMS metabolomics experiments suggested that the G6PD substrate (G6P) did not accumulate to high levels during PGD inhibition (Fig. 2a). This finding contrasted with our previous observations for 6PG [13], and raised the intriguing possibility that PGD might not operate entirely within the oxPPP in distant metastases. We therefore examined other glucose-derived metabolites by LC–HRMS in order to identify alternative reactions that might lie upstream of PGD, which we hypothesized would manifest as large elevations in signal during PGD inhibition (similar to 6PG). This analysis identified three structurally-related uronic-aldonic acids that showed striking elevations (Fig. 2a). Remarkably, these metabolites could be ordered into a linear series of reactions that connected glucose to 6PG outside the PPP, which strongly resembled a core pentose conversion pathway (PCP; Fig. 2b). We therefore tested the possibility that a PCP was selected in distant metastases to stimulate high PGD catalytic activity and support PGD-dependent tumorigenesis.
Fig. 2.
PCP substrates stimulate PGD catalytic activity. a Steady state LC–HRMS signals were measured for the metabolites indicated, from 38Lg cells treated with DMSO or 250 μM 6AN for 3 days. Unlike G6P/F6P, levels of glucuronate/gulonate/gluconate showed large elevations in response to 6AN, similar to what we previously observed for 6PG. Structures of the elevated pentoses are depicted in the boxed area, along with 6PG. b The PCP model predicts that distant metastases can bypass the oxPPP to manufacture 6PG and activate PGD. Step 1: Glucose is shunted to UDP-Glucose-6-Dehydrogenase (UGDH), which generates UDP-glucuronate. A host of enzymes can then catalyze the hydrolysis of UDP-glucuronate to glucuronate. Step 2: Glucuronate is converted to gulonate by Aldo-Keto Reductase 1A1 (AKR1A1). Step 3: Gulonate is converted to gluconate. The absence of a functional gulonolactone oxidase in humans prevents gulonate or gluconate from exiting the PCP and entering into the ascorbic acid biosynthetic pathway. Step 4: Gluconate is phosphorylated to 6PG by Gluconokinase (IDNK), which requires ATP. Step 5: PGD catalysis consumes the 6PG to produce NADPH, which is measured by PGD enzyme assays. c Enzyme assays were conducted by incubating 50 μg of cell extracts with 0.2 mM NADP, 0.8 mM of the indicated PCP substrates, and 2 mM ATP. PCP metabolites selectively stimulated PGD activity in liver and lung metastases but not control cells. d Enzyme assays were performed on the lung metastasis from panel c, for the indicated conditions. Reactions failed to proceed in the absence of ATP (“No ATP”), and pre-incubating extracts with ATP for 1 h at 37 °C prior to addition of NADP (“ATP Primed”) resulted in a sharp initial acceleration in NADPH production upon addition of NADP, which then settled back to steady state rates (“Steady State”). e RNAi (siRNA, shRNA) against PGD blocked the ability of PCP enzymes to stimulate NADPH production in enzyme assays, confirming that the upstream PCP reactions were PGD-dependent. For all experiments in this figure, statistical significance was calculated using 2-tailed t-tests (*p < 0.05)
We first asked whether the PCP substrates might selectively maintain high PGD catalysis in distant metastases, as predicted if these are converted to 6PG upstream of the PGD reaction. To address this possibility in-depth, we performed a series of PGD enzyme assays on cell extracts harvested from PGD-dependent liver and lung metastases and control extracts harvested from a PGD-independent peritoneal metastasis from the same patient. Immortalized, non-malignant human pancreatic ductal epithelial cells were also included as an additional control. Strikingly, each PCP substrate was able to substitute for 6PG in enzyme assays and selectively produce NADPH in liver and lung metastases (Fig. 2c) over a range of concentrations (Supplementary Fig. 2a), as predicted if the PCP activates PGD-driven NADPH production (Fig. 2b). This was dependent on the presence of ATP (Fig. 2d), which is also predicted since step 4 of the pathway requires a gluconate kinase (Fig. 2b). Another prediction is that pre-incubating extracts with PCP substrates and ATP prior to PGD activation (with NADP) would accelerate reaction rates, since flux of each substrate would be drawn forward into the pathway resulting in 6PG accumulation (Supplementary Fig. 2b). Consistent with this, pre-incubation of extracts with each PCP substrate and ATP (“ATP primed”), followed by activation of PGD with NADP, strongly accelerated NADPH production relative to steady state rates (Fig. 2d, Supplementary Fig. 2c). Finally, both acute (siRNA) and stable (shRNA) knockdown of PGD blocked NADPH production from PCP substrates with comparable efficacy as 6PG itself (Fig. 2e), confirming that PCP-driven NADPH production reflected the activation of PGD. Thus, each pentose substrate selectively activated PGD catalysis according to predictions of the PCP model.
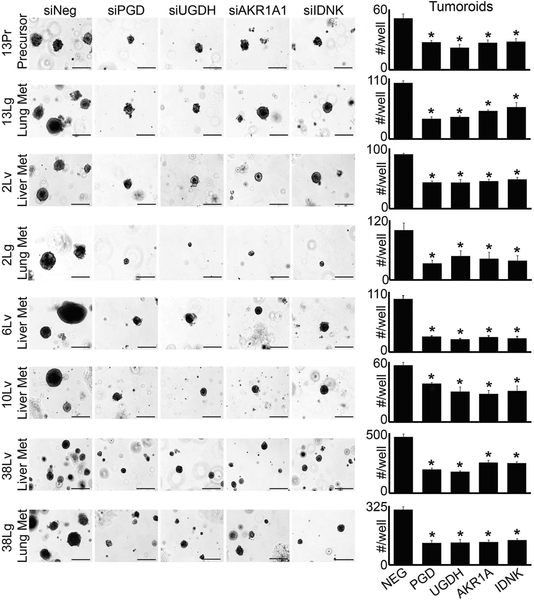
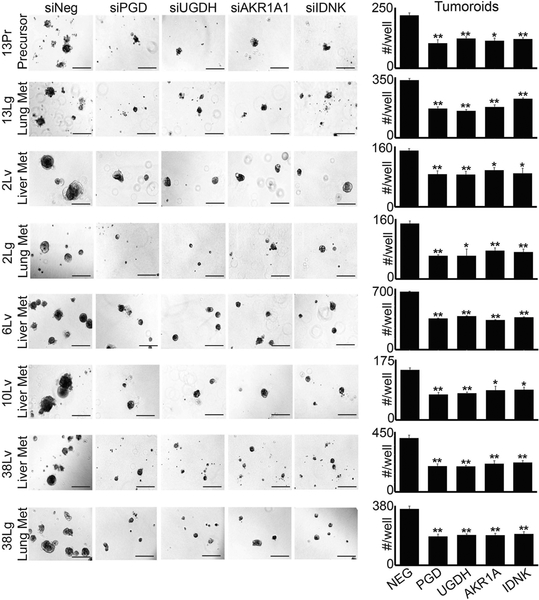
Specific enzymes have been reported to catalyze forward reactions [51–53] that are predicted to lie within the PCP (Fig. 2b). We therefore tested whether these enzymes were required to selectively maintain tumorigenic capacity in distant metastases, as would be expected if they function upstream of PGD. Remarkably, RNAi knockdown of each individual PCP enzyme (UGDH, AKR1A1, and IDNK) recurrently impaired 3D tumorigenesis across distant metastatic subclones with comparable efficacy as PGD knockdown, which we replicated across three separate 3D tumor forming assays (Figs. 3 and 4; Supplementary Fig. 3). In contrast, we observed minimal effects on 3D tumorigenesis across a panel of PGD-independent control cells, despite efficient knockdowns (Supplementary Fig. 4). Thus, each enzyme predicted to lie within the PCP was required to support the tumorigenic capacity of distant metastatic subclones, similar to PGD.
Fig. 3.
PCP enzymes are required for 3D tumorigenesis (Matrigel embedding assays). Cells isolated from the distant metastatic subclones indicated were transfected with non-targeting control siRNAs (siNeg) or siRNAs against the indicated genes (Sigma), trypsinized into single cells, and 1000 cells were embedded in 50 μl of Matrigel discs in triplicate, similar to conventional organoid assays [54] using DMEM with 10% FBS and no additional supplements. Tumoroids were photographed and counted after 3 weeks of tumoroid growth. Knockdown of each individual PCP enzyme impaired tumoroid growth relative to siNeg controls, similar to PGD (see Supplementary Fig. 4 for knockdown efficacies). Statistical significance was calculated using 2-tailed t-tests (*p < 0.05)
Fig. 4.
PCP enzymes are required for 3D tumorigenesis (suspension Matrigel assays). Cells isolated from the distant metastatic subclones indicated were transfected with non-targeting control siRNAs (siNeg) or siRNAs against the indicated genes (Sigma), trypsinized into single cells, and 4000 cells/well were plated in suspension Matrigel assays as previously described (Fig. 1). Similar to embedding assays, knockdown of each PCP enzyme impaired tumoroid growth relative to siNeg controls. Statistical significance was calculated using 2-tailed t-tests (*p < 0.05; **p ≤ 0.001)
Collectively, our results define a metastasis-intrinsic series of pentose conversion reactions that support PGD-dependent tumorigenesis. We hypothesize that these reactions were selected during the evolution of distant metastasis to manufacture PGD substrate outside of the rate-limiting oxPPP (Fig. 2b), thereby enabling high PGD catalytic activity. We caution that pleiotropic effects of the upstream PCP components (glucuronate) or unknown functions of the sparsely-studied downstream components (gulonate and gluconate) could also influence processes outside of the PCP. For example, UGDH catalyzes the formation of UDP-glucuronate from UDP-glucose. A large number of downstream enzymes are able to hydrolyze UDP-glucuronate to glucuronate, which can then be used for proteoglycan synthesis or glucuronidation reactions [51] outside the PCP. However, glucuronate that does not enter these other pathways (or is salvaged from them) is converted to gulonate by AKR1A1 [51, 52]. Gulonate and gluconate may be especially suited to enter the PCP (Fig. 2b), since humans have lost the ability to divert the lactone forms of these pentoses into the ascorbic acid biosynthetic pathway [52]. We also predict that an enzymatic activity is required to convert gulonate into gluconate, which we are pursuing in other studies. Finally, it remains a formal possibility that glucuronate or gulonate could also serve as direct precursors of 6PG, which would require additional glucuronate or gulonate kinase activities, similar to the gluconate kinase activity of IDNK.
Primary PDACs rely on genetically-encoded metabolic adaptations to support tumorigenesis under hypoglycemic conditions, as found within their dense stromal micro-environment. Our findings raise the possibility that distant metastases evolve additional metabolic adaptations to support tumorigenesis under glucose-replete conditions, as found along the metastatic route. The PCP could represent one of many such adaptations, and it is possible that regulatory inputs from other pathways may influence flux through the core PCP itself. Because distant metastases are strongly PGD-dependent, targeting unique adaptations such as the PCP that enable and/or reward this vulnerability could represent an effective therapeutic strategy against metastatic PDAC, which remains one of the most lethal of all human malignancies.
Supplementary Material
Acknowledgments
This work was supported by the AACR Pancreatic Cancer Action Network Pathway to Leadership grant (OGM), the Vanderbilt GI SPORE (OGM), and the Vanderbilt Digestive Diseases Research Center (OGM).
Footnotes
Electronic supplementary material The online version of this article (https://doi.org/10.1038/s41388-018-0346-5) contains supplementary material, which is available to authorized users.
Conflict of interest The authors declare that they have no conflict of interest.
References
- 1.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–21. [DOI] [PubMed] [Google Scholar]
- 2.Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. N Engl J Med. 2014;371:1039–49. [DOI] [PubMed] [Google Scholar]
- 3.Vincent A, Herman J, Schulick R, Hruban RH, Goggins M. Pancreatic cancer. Lancet. 2011;378:607–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Makohon-Moore A, Iacobuzio-Donahue CA. Pancreatic cancer biology and genetics from an evolutionary perspective. Nat Rev Cancer. 2016;16:553–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yachida S, White CM, Naito Y, Zhong Y, Brosnan JA, Macgregor-Das AM, et al. Clinical significance of the genetic landscape of pancreatic cancer and implications for identification of potential long-term survivors. Clin Cancer Res. 2012;18:6339–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Embuscado EE, Laheru D, Ricci F, Yun KJ, de Boom Witzel S, Seigel A, et al. Immortalizing the complexity of cancer metastasis: genetic features of lethal metastatic pancreatic cancer obtained from rapid autopsy. Cancer Biol Ther. 2005;4:548–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iacobuzio-Donahue CA, Fu B, Yachida S, Luo M, Abe H, Henderson CM, et al. DPC4 gene status of the primary carcinoma correlates with patterns of failure in patients with pancreatic cancer. J Clin Oncol. 2009;27:1806–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vanharanta S, Massague J. Origins of metastatic traits. Cancer Cell. 2013;24:410–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haeno H, Gonen M, Davis MB, Herman JM, Iacobuzio-Donahue CA, Michor F. Computational modeling of pancreatic cancer reveals kinetics of metastasis suggesting optimum treatment strategies. Cell. 2012;148:362–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rhim AD, Mirek ET, Aiello NM, Maitra A, Bailey JM, McAllister F, et al. EMT and dissemination precede pancreatic tumor formation. Cell. 2012;148:349–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yachida S, Jones S, Bozic I, Antal T, Leary R, Fu B, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McDonald OG, Li X, Saunders T, Tryggvadottir R, Mentch SJ, Warmoes MO, et al. Epigenomic reprogramming during pancreatic cancer progression links anabolic glucose metabolism to distant metastasis. Nat Genet. 2017;49:367–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carter H, Samayoa J, Hruban RH, Karchin R. Prioritization of driver mutations in pancreatic cancer using cancer-specific high-throughput annotation of somatic mutations (CHASM). Cancer Biol Ther. 2010;10:582–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Makohon-Moore AP, Zhang M, Reiter JG, Bozic I, Allen B, Kundu D, et al. Limited heterogeneity of known driver gene mutations among the metastases of individual patients with pancreatic cancer. Nat Genet. 2017;49:358–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murphy SJ, Hart SN, Lima JF, Kipp BR, Klebig M, Winters JL, et al. Genetic alterations associated with progression from pancreatic intraepithelial neoplasia to invasive pancreatic tumor. Gastroenterology. 2013;145:1098–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Campbell PJ, Yachida S, Mudie LJ, Stephens PJ, Pleasance ED, Stebbings LA, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murphy SJ, Hart SN, Halling GC, Johnson SH, Smadbeck JB, Drucker T, et al. Integrated genomic analysis of pancreatic ductal adenocarcinomas reveals genomic rearrangement events as significant drivers of disease. Cancer Res. 2016;76:749–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Notta F, Chan-Seng-Yue M, Lemire M, Li Y, Wilson GW, Connor AA, et al. A renewed model of pancreatic cancer evolution based on genomic rearrangement patterns. Nature. 2016;538:378–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morton JP, Timpson P, Karim SA, Ridgway RA, Athineos D, Doyle B, et al. Mutant p53 drives metastasis and overcomes growth arrest/senescence in pancreatic cancer. Proc Natl Acad Sci USA. 2010;107:246–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mueller S, Engleitner T, Maresch R, Zukowska M, Lange S, Kaltenbacher T, et al. Evolutionary routes and KRAS dosage define pancreatic cancer phenotypes. Nature. 2018;554:62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qiu W, Sahin F, Iacobuzio-Donahue CA, Garcia-Carracedo D, Wang WM, Kuo CY, et al. Disruption of p16 and activation of Kras in pancreas increase ductal adenocarcinoma formation and metastasis in vivo. Oncotarget. 2011;2:862–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhong Y, Macgregor-Das A, Saunders T, Whittle MC, Makohon-Moore A, Kohutek ZA, et al. Mutant p53 together with TGFbeta signaling influence organ-specific hematogenous colonization patterns of pancreatic cancer. Clin Cancer Res. 2016;23:1607–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bryant KL, Mancias JD, Kimmelman AC, Der CJ. KRAS: feeding pancreatic cancer proliferation. Trends Biochem Sci. 2014;39:91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Commisso C, Davidson SM, Soydaner-Azeloglu RG, Parker SJ, Kamphorst JJ, Hackett S, et al. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature. 2013;497:633–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davidson SM, Jonas O, Keibler MA, Hou HW, Luengo A, Mayers JR, et al. Direct evidence for cancer-cell-autonomous extracellular protein catabolism in pancreatic tumors. Nat Med. 2016;23:235–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeNicola GM, Karreth FA, Humpton TJ, Gopinathan A, Wei C, Frese K, et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475:106–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Halbrook CJ, Lyssiotis CA. Employing metabolism to improve the diagnosis and treatment of pancreatic cancer. Cancer Cell. 2017;31:5–19. [DOI] [PubMed] [Google Scholar]
- 30.Kamphorst JJ, Cross JR, Fan J, de Stanchina E, Mathew R, White EP, et al. Hypoxic and Ras-transformed cells support growth by scavenging unsaturated fatty acids from lysophospholipids. Proc Natl Acad Sci USA. 2013;110:8882–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kamphorst JJ, Nofal M, Commisso C, Hackett SR, Lu W, Grabocka E, et al. Human pancreatic cancer tumors are nutrient poor and tumor cells actively scavenge extracellular protein. Cancer Res. 2015;75:544–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rhim AD, Oberstein PE, Thomas DH, Mirek ET, Palermo CF, Sastra SA, et al. Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer Cell. 2014;25:735–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosenfeldt MT, O’Prey J, Morton JP, Nixon C, MacKay G, Mrowinska A, et al. p53 status determines the role of autophagy in pancreatic tumour development. Nature. 2013;504:296–300. [DOI] [PubMed] [Google Scholar]
- 34.Son J, Lyssiotis CA, Ying H, Wang X, Hua S, Ligorio M, et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 2013;496:101–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sousa CM, Biancur DE, Wang X, Halbrook CJ, Sherman MH, Zhang L, et al. Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature. 2016;536:479–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ying H, Kimmelman AC, Lyssiotis CA, Hua S, Chu GC, Fletcher-Sananikone E, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149:656–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kohler E, Barrach H, Neubert D. Inhibition of NADP dependent oxidoreductases by the 6-aminonicotinamide analogue of NADP. FEBS Lett. 1970;6:225–8. [DOI] [PubMed] [Google Scholar]
- 39.Boj SF, Hwang CI, Baker LA, Chio II, Engle DD, Corbo V, et al. Organoid models of human and mouse ductal pancreatic cancer. Cell. 2015;160:324–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheung WK, Zhao M, Liu Z, Stevens LE, Cao PD, Fang JE, et al. Control of alveolar differentiation by the lineage transcription factors GATA6 and HOPX inhibits lung adenocarcinoma metastasis. Cancer Cell. 2013;23:725–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ridky TW, Chow JM, Wong DJ, Khavari PA. Invasive three-dimensional organotypic neoplasia from multiple normal human epithelia. Nat Med. 2010;16:1450–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi J, Wang E, Milazzo JP, Wang Z, Kinney JB, Vakoc CR. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat Biotechnol. 2015;33:661–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dyson JE, D’Orazio RE. 6-Phosphogluconate dehydrogenase from sheep liver: inhibition of the catalytic activity by fructose-1,6-diphosphate. Biochem Biophys Res Commun. 1971;43:183–8. [DOI] [PubMed] [Google Scholar]
- 44.Hanau S, d’Empaire LP, Capone I, Alberighi S, Montioli R, Dallocchio F. Evidence for dimer/tetramer equilibrium in Trypanosoma brucei 6-phosphogluconate dehydrogenase. Biochim Biophys Acta. 2013;1834:2647–52. [DOI] [PubMed] [Google Scholar]
- 45.Hitosugi T, Zhou L, Elf S, Fan J, Kang HB, Seo JH, et al. Phosphoglycerate mutase 1 coordinates glycolysis and biosynthesis to promote tumor growth. Cancer Cell. 2012;22:585–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rippa M, Giovannini PP, Barrett MP, Dallocchio F, Hanau S. 6-Phosphogluconate dehydrogenase: the mechanism of action investigated by a comparison of the enzyme from different species. Biochim Biophys Acta. 1998;1429:83–92. [DOI] [PubMed] [Google Scholar]
- 47.Shan C, Elf S, Ji Q, Kang HB, Zhou L, Hitosugi T, et al. Lysine acetylation activates 6-phosphogluconate dehydrogenase to promote tumor growth. Mol Cell. 2014;55:552–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singh A, Happel C, Manna SK, Acquaah-Mensah G, Carrerero J, Kumar S, et al. Transcription factor NRF2 regulates miR-1 and miR-206 to drive tumorigenesis. J Clin Invest. 2013;123:2921–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yao P, Sun H, Xu C, Chen T, Zou B, Jiang P, et al. Evidence for a direct crosstalk between malic enzyme and the pentose phosphate pathway via structural interactions. J Biol Chem. 2017;292:17113–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Patra KC, Hay N. The pentose phosphate pathway and cancer. Trends Biochem Sci. 2014;39:347–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Egger S, Chaikuad A, Kavanagh KL, Oppermann U, Nidetzky B. UDP-glucose dehydrogenase: structure and function of a potential drug target. Biochem Soc Trans. 2010;38:1378–85. [DOI] [PubMed] [Google Scholar]
- 52.Linster CL, Van Schaftingen E. Vitamin C. Biosynthesis, recycling and degradation in mammals. FEBS J. 2007;274:1–22. [DOI] [PubMed] [Google Scholar]
- 53.Rohatgi N, Nielsen TK, Bjorn SP, Axelsson I, Paglia G, Voldborg BG, et al. Biochemical characterization of human gluconokinase and the proposed metabolic impact of gluconic acid as determined by constraint-based metabolic network analysis. PLoS One. 2014;9:e98760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.