Key Points
Pembrolizumab induced a complete remission in a patient with high-risk smoldering multiple myeloma who also had high-risk myeloma features.
Immune and transcriptomic analyses identified a distinct profile that indicated a preexisting immune response in the responder.
Abstract
Multiple myeloma is, in most patients, an incurable cancer. Its precursors can be identified with routine tests setting the stage for early intervention to prevent active myeloma. We investigated the efficacy and safety of pembrolizumab, an antiprogrammed cell death 1 antibody, in smoldering myeloma patients with intermediate/high risk of progression to symptomatic myeloma. Thirteen patients were treated with a median number of 8 cycles. One patient achieved a stringent complete response with bone marrow next-generation sequencing negativity at 10−4 that is ongoing at 27 months (8%); 11 had stable disease (85%), and 1 progressed (8%). Three patients discontinued therapy due to immune-related adverse events: 2 with transaminitis and 1 due to tubulointerstitial nephritis. Immune profiling of bone marrow samples at baseline showed markers associated with a preexisting immune response in the responder compared with nonresponders and features of increased T-cell exhaustion in nonresponders. Consistent with this, transcriptome sequencing of bone marrow samples at baseline revealed an increased interferon-γ signature in the responder compared with the nonresponders. In summary, our results suggest that smoldering myeloma may be immunogenic in a subset of patients, and therapies that enhance antitumor T-cell responses may be effective in preventing its progression. This trial was registered at www.clinicaltrials.gov as #NCT02603887.
Visual Abstract
Introduction
The current standard for management of patients with smoldering multiple myeloma (SMM) is watchful waiting. However, a subset of these patients is at significant risk of progression to symptomatic multiple myeloma (MM). Expression of programmed cell death 1 ligand 1 (PD-L1) on tumor cells was associated with increased risk of progression from SMM to MM, suggesting a role for immune checkpoint blockade to prevent development of symptomatic MM.1,2 In addition, expression of PD-L1 on tumor cells as well as stromal cells and PD-1 on CD4+ and CD8+ T cells has been reported in MM.3-6 Pembrolizumab is a humanized immunoglobulin G4 monoclonal antibody against programmed cell death-1 (PD-1) receptor that blocks the interaction between PD-1 and PD-L1 and is used for treatment of solid tumors and lymphoma.7 Here, we report the clinical and correlative results of a pilot study of pembrolizumab in intermediate- and high-risk SMM patients (I-HR-SMM).
Methods
This is a pilot single-institution study of pembrolizumab monotherapy in I-HR-SMM (supplemental Tables 1 and 2). The trial was conducted by the Simon’s minimax design; 12 patients were to be enrolled in the first stage, and if at least 1 patient had ≥partial response after 8 cycles of therapy, 4 more patients were to be enrolled for a total of 16 patients. Pembrolizumab 200 mg was administered as a 30-minute IV infusion every 21 days for up to 8 cycles, with an option to continue up to 24 cycles if there was continued clinical benefit (≥minor response) and without serious adverse events (AEs) after 8 cycles.
The primary objective was to determine the overall response rate (ORR) (partial response or better) after 8 cycles of treatment per International Myeloma Working Group criteria for MM.8 Secondary objectives were to evaluate clinical benefit rate (minor response or better) after 8 cycles of treatment, safety and tolerability, and duration of response. Exploratory objectives included the rate of minimal residual disease (MRD) negativity at complete remission (CR) by Clonoseq (Adaptive Biotechnologies, Seattle, WA), molecular profiling (including whole-exome sequencing [WES] and gene expression profiling [GEP]) at baseline and progression, immunophenotypic characterization evaluated by flow cytometry on bone marrow aspirates, and/or peripheral blood collected at baseline and at the end of treatment.
AEs were graded according to the National Cancer Institute Common Terminology Criteria for Adverse Events (NCI-CTCAE ver. 4). Safety and toxicity were evaluated based on changes in physical examination, vital signs, and clinical laboratory and report of any AEs, including laboratory abnormalities, that occurred from cycle 1 day 1 until 30 days after the last dose of study agent and severe AEs (including laboratory abnormalities) occurring after 30 days if considered related to study drug.
Cytogenetics and fluorescence in situ hybridization (FISH) were performed at baseline. Serial peripheral blood mononuclear cells and bone marrow aspirate samples were collected for comprehensive immunophenotyping using multiparametric flow cytometry and molecular profiling with GEP70 and WES of bulk tumor cells DNA as well as RNA sequencing of both tumor and microenvironment cells from bulk RNA.
To study the effect of pembrolizumab on the frequency and functional status of various immune cell subsets, we performed comprehensive immunophenotyping of immune activation/inhibitory cell markers in paired bone marrow aspirate patient samples at baseline and at end of treatment. Markers analyzed include T-cell activating receptors (CD134, CD137, CD40L, ICOS, GITR), natural killer (NK) cell-activating receptors (NKG2D, NKp46, NKp44, NKp30, CD226), NK cell activating ligands (MICA/B, ULBP1, ULBP2, ULBP3, CD112, CD155), T/NK inhibitory receptors (PD1, LAG3, TIM3, CD244, CD160, BTLA, CD200R, CTLA4, TIGIT), NK cell inhibitory receptors (KIR2DL1/L2/L3/S1/S2), T/NK inhibitory ligands (PD-L1, PD-L2, B7-H3), and markers of immune cell subsets, including naive, effector, and memory T cells, regulatory T cells, NK cells, macrophages, and myeloid-derived suppressor cells. We designed 11 tubes with antibodies to the above-mentioned markers. After red blood cell removal, bone marrow cells were distributed in 11 tubes and stained for 30 minutes on ice before being fixed with either FoxP3 staining buffer or 1% paraformaldehyde. Tubes fixed with FoxP3 staining buffer were further stained for FoxP3, and all samples were acquired using a flow cytometer. Samples were acquired using a BD LSR Fortessa (BD Biosciences) and analyzed by FlowJo (TreeStar).
The protocol was approved by the Institutional Review Board of The University of Texas MD Anderson Cancer Center and the US Food and Drug Administration (FDA) in accordance with the Declaration of Helsinki and the guidelines for Good Clinical Practice. The trial was registered with www.clinicaltrials.gov, identifier #NCT02603887.
Additional methods on genomic profiling are included in the supplemental Material.
Results
Patients
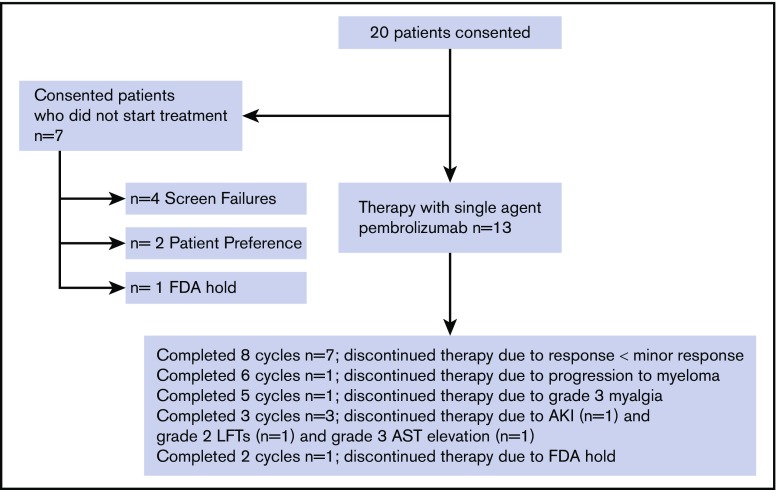
Twenty patients were screened, and 13 patients (12 on stage 1 and 1 on stage 2) were treated at MD Anderson Cancer Center between 16 August 2016 and 22 August 2017. Seven patients who consented were not treated due to the following: 3 patients had a diagnosis of MM and went on to receive standard myeloma treatment; 1 patient had a diagnosis of monoclonal gammopathy of unknown significance and proceeded to watchful waiting; 2 patients withdrew consent prior to treatment due to personal preference (distance to treatment center); and 1 patient was going to be treated but the study was placed on hold by the FDA9 (Figure 1). Demographic and baseline characteristics, including risk model classification for the treated patients on the study, are summarized in supplemental Table 2. Twelve (12/13; 92%) patients had high-risk SMM by at least one of the SMM risk stratification criteria. Several patients were classified as either low, intermediate, or high risk by different models, underscoring the need for a standardized model (supplemental Table 2).
Figure 1.
Consort diagram for pilot study of pembrolizumab therapy in smoldering myeloma. AKI, acute kidney injury attributed to pembrolizumab; AST, aspartate aminotransferase elevation attributed to pembrolizumab; LFTs, liver function tests elevation attributed to pembrolizumab.
Efficacy and tolerability
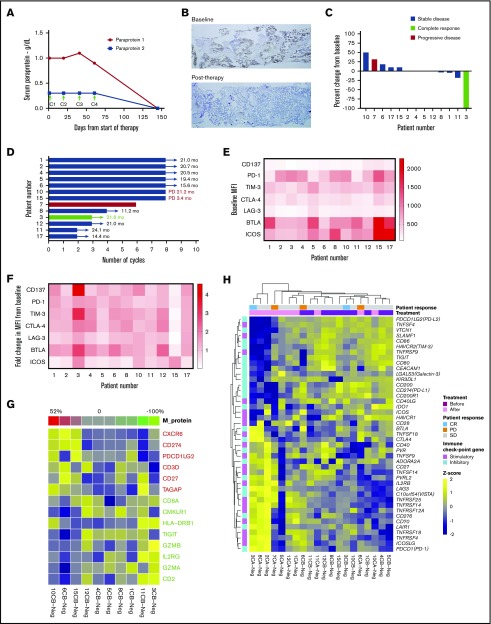
All 13 treated patients completed at least 1 cycle of therapy (median 8, range 2-8 cycles) and were evaluable for safety and efficacy (Figure 1). The ORR and clinical benefit rate at or before 8 cycles of therapy were both 7.7% (1/13 patients). One patient achieved a stringent complete remission (sCR; Clonoseq next-generation sequencing [NGS] negative at 10−4) after 3 cycles of therapy (7.7%), 11 patients had stable disease (SD) at ≤8 cycles (84.6%), and 1 patient progressed to MM requiring treatment after 6 cycles of therapy (7.7%) (Figure 2A-D). Seven (53.8%) patients completed 8 cycles of therapy, and their response after 8 cycles was SD (Figure 2D).
Figure 2.
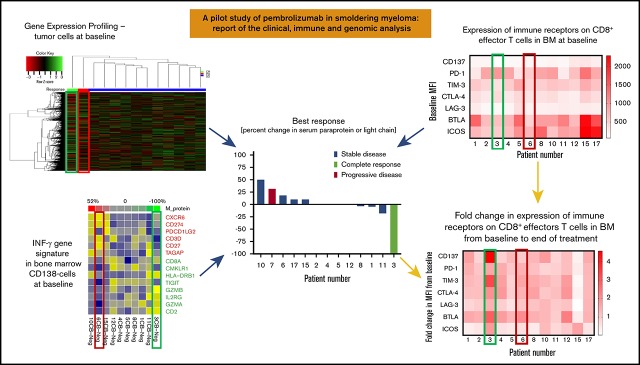
Clinical outcomes and correlative analyses in patients with smoldering myeloma treated with pembrolizumab. (A) Paraprotein 1 and 2 levels in the serum in the extraordinary responder (patient 3) at baseline and after start of pembrolizumab therapy. Arrows indicate day 1 of start of cycles 1, 2, 3, and 4 (C1, C2, C3, C4). Treatment was stopped after C3 due to transaminitis. (B) Immunohistochemistry for plasma cells was performed on bone marrow samples at baseline and after therapy with pembrolizumab using CD138 stain. Baseline sample showed 50% infiltration of plasma cells, and posttherapy sample showed 1% to 2% plasma cells, all of which were of normal phenotype by flow cytometry (data not shown). Original magnification ×5. (C) Best response during treatment period as determined by percent change in serum paraprotein or light chain (in light chain only patients) levels from baseline in all 13 patients. (D) Duration of response and/or clinical benefit in all 13 patients calculated from day 1 of last cycle of pembrolizumab. Number of months of ongoing response, SD, or progressive disease is shown for each patient. (E) Mean fluorescence intensity (MFI) values for expression of various immune stimulatory (CD137 and ICOS) and inhibitory (PD-1, TIM-3, CTLA-4, LAG-3, and BTLA) receptors on CD8+ effectors T cells in bone marrow samples available at baseline from 12 patients are shown as a heat map. (F) Fold change in MFI for the receptors on CD8+ effectors T cells in bone marrow samples from baseline to end of treatment period is shown as a heat map (N = 12 patients). (G) Correlation between IFN-γ gene signature in the CD138− cells in the bone marrow samples at baseline and percent change in paraprotein level (or light chain in light chain–only patients) in serum with therapy. (H) Heat map showing GEP of a curated list of immune stimulatory and immune checkpoint genes in CD138− (Neg) cells from bone marrow samples obtained from patients before (B) and/or after (A) treatment with pembrolizumab.
One patient achieved an sCR after 3 cycles of therapy with Clonoseq NGS negativity at a level of 10−4 and MRD indeterminate at 10−6. This patient had high-risk SMM by GEP70 (score 44.9, subtype LB [low bone disease]) and FISH (deletion 17p and amplification of cyclin-dependent kinase regulatory subunit 1 [CKS1B]) and 50% plasma cells infiltration in the bone marrow core biopsy at study entry (Figure 2B).
One patient progressed to MM requiring therapy 14 weeks after completion of the eighth cycle of therapy. The patient was started on treatment with carfilzomib, lenalidomide, and dexamethasone. After 4 cycles of carfilzomib, lenalidomide, and dexamethasone, while in very good partial response, the patient suffered cardiac arrest presumably due to pulmonary embolus (the family declined an autopsy). The patient had been taking aspirin 325 mg orally daily as thromboprophylaxis. A second patient progressed to MM 21 months after the last dose of pembrolizumab was given. No other patient progressed to MM at the time of submission of this report (Figure 2D). The median progression-free survival (PFS) and overall survival have not been reached (supplemental Figure 1). The median follow-up time on the study for the remaining 12 censored observations was 24.16 months (15.80 to 26.22 months).
Three patients discontinued therapy due to immune-related AEs: grade 3 transaminitis (n = 1), grade 2 transaminitis without resolution within 12 weeks (n = 1), and grade 3 acute kidney injury due to tubulo-interstitial nephritis (n = 1) (supplemental Table 3). Transaminitis resolved with discontinuation of treatment in both patients, and acute kidney injury resolved with corticosteroid administration. A fourth patient discontinued therapy due to grade 3 myalgia that was determined as not an immune-related AE. There were no grade 4/5 AEs. The clinical trial was stopped after the last patient completed 2 cycles of therapy per FDA guidance due to unexpected deaths in myeloma patients observed in other clinical trials with pembrolizumab.10
Flow cytometry analysis
Twelve (12/13; 92%) patients had baseline and end-of-study bone marrow aspirate samples available for this analysis. Immune profiling of bone marrow samples by multiparametric flow cytometry showed that the extraordinary responder (patient 3) had high levels of PD-1 expression and low levels of other inhibitory receptors, such as TIM-3, LAG-3, and BTLA on CD8+ T cells compared with nonresponders at baseline (Figure 2E). Interestingly, the expression of various stimulatory and inhibitory receptors, such as CD137 (4-1BB), PD-1, TIM-3, CTLA-4, LAG-3, BTLA, and ICOS, increased markedly on CD8+ T cells in the bone marrow after therapy (Figure 2F).
GEP analysis
Baseline GEP70 analysis for CD138+ cells from bone marrow aspirates was available for all patients. After running different clustering algorithms, we found that the patient in CR almost always clustered separately from the other response groups. In addition, the patient who progressed to MM while on treatment tended to cluster away from the CR patient and the other patients who had SD. This pattern was consistent for multiple clustering methods, including average linkage (Euclidean), average linkage (correlation), single linkage (Euclidean), single linkage (correlation), and complete linkage (Euclidean). The heat map was drawn based on the complete lineage method for both genes and patients (supplemental Figure 2). Probes for X inactive-specific transcript (XIST) and SLIT and NTRK-like family member 1 (SLITRK4) ranked among the top 10 in enriched probe comparisons between the CR patient and the patient who progressed to MM (PD) or the patients who had SD as best response. Pathways like Ephrin receptor signaling, integrin signaling, and Tec kinase signaling were ranked among the top 20 enriched pathways in comparison between CR and SD.
The genes XIST and SLITRK4 were highly expressed in the responder compared with the rest of the patients. XIST is a long noncoding RNA known for its role in X chromosome inactivation in mammals and is associated with solid tumors.11 To our knowledge, its role in myeloma has not been described. SLITRK4 encodes a transmembrane protein of the SLITRK family highly expressed in the brain and adrenal glands and suppresses neurite growth. To our knowledge, high expression of SLITKR4 has not been described before in myeloma.
Genomic analysis: DNA and RNA sequencing of plasma cells and microenvironment
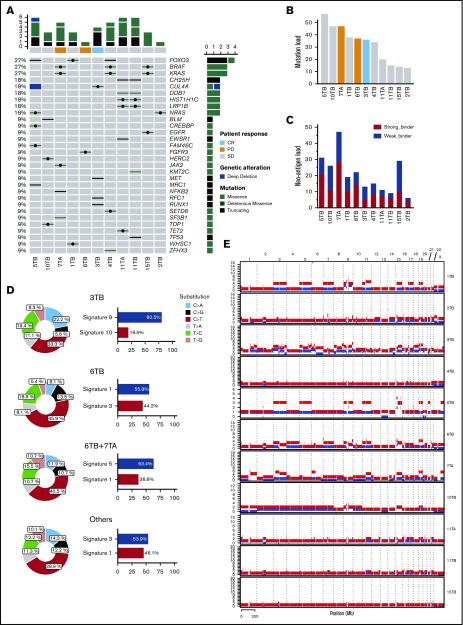
WES of CD138+ plasma cells was performed for 9 (9/13; 70%) patients at baseline and 2 (2/13; 15%) patients at end of study, with matched germline DNA from peripheral blood mononuclear cells as control. The most commonly mutated genes at a sample level either at baseline or end of treatment (3/9; 33%) were FOXO3 (27%), BRAF (27%), and KRAS (27%), (Figure 3A). The total genetic burden resulting from accumulated nonsynonymous somatic mutations (mutational load) ranged from 13 to 57 (32.5 ± 15.1) (Figure 3B). Neoantigen analysis was also performed showing that most patients have a neoantigen load exceeding 10 neoantigens (Figure 3C). Three base-substitution signatures in specimens from either CR or PD patients (Figure 3D) were identified according to COSMIC nomenclature.12 The predominant mutational signatures included signature 9 (81%, 3TB [tumor baseline], CR), which has been found in chronic lymphocytic leukemias and malignant B-cell lymphomas, signature 1 (56%, 6TB, PD), which is associated with a clocklike mutational process and a higher cell turnover rate,13 signature 3 (44%, 6TB, PD), which is associated with failure of DNA double-strand break-repair by homologous recombination, and signature 5 (63%, 6TB and 7TA [tumor after treatment] merged, PD), which is uncharacterized.
Figure 3.
Genomic studies in trial patients. Mutational profile (A), mutational load (B), neoantigen load (C), mutational signatures (D), and copy number alterations (E) in bone marrow samples obtained at baseline from patients with smoldering myeloma.
Mutational signatures have been described in a small series of 11 SMM patients with matched progression samples.14 Both this study and ours describe signatures 1, 5, and 9. We additionally describe signature 3 in the patient who progressed to MM while on study, which is associated with DNA repair failure and to our knowledge has not been described before in SMM.
Frequent copy number alterations included copy number gains on chr3, chr5, chr9, chr11, 15q, and chr19 (Figure 3E). The CR patient (3TB) had a higher degree of aneuploidy, particularly the events of copy number gains and copy number alterations, that covered not only odd-numbered chromosomes but also even-numbered chromosomes.
Whole transcriptome sequencing (RNA-seq) was performed on 31 bone marrow specimens (either CD138+ or CD138−) isolated from 12 patients either at baseline or after treatment. GEP against a curated list of immune checkpoint genes15 demonstrated strong activation of immune stimulatory genes (n = 15) after treatment (Figure 2H), notably in patient 3, the sCR patient. The expression levels of inhibitory immune checkpoint genes, such as CD274 (PD-L1), PDCD1LG2 (PD-L2), HAVCR2 (TIM-3), and IDO1, were significant decreased in posttreatment samples compared with samples at baseline (false discovery rate [FDR] q value = 0.04, fold change = −1.4). However, in the posttreatment sample of patient 6, the progressor, expression of inhibitory immune checkpoint genes, including CTLA-4, PD-1, IDO1, and TIM-3, was dramatically increased.
In addition to immune checkpoint genes in CD138− cells, we performed an unbiased analysis to identify differentially expressed genes (DEGs) in CD138+ tumor cells between the PD (n = 1, 6RB) and SD (n = 4) patients. A total of 649 protein-coding genes showed differential expression between these 2 groups, with a fold change of ≥2 or ≤ −2, and the FDR q value ≤ 0.05. Among the 649 DEGs, 106 genes were upregulated in the PD baseline sample (supplemental Figure 3A). Gene set enrichment analysis of these 106 genes suggested a significant enrichment of genes in the oxidative phosphorylation pathway (FDR q value = 0, normalized enrichment score [NES] = 2.7) as well as MTORC1 signaling (FDR q value = 0.02, NES = 1.8) (supplemental Figure 3B). The top upregulated DEGs included FGFR3, WHSC1, and MAF, which were reported to be associated with t4,14 translocation in MM.16
We also compared baseline tumor microenvironment samples (CD138− cells) between the SD (n = 9) and CR (n = 1, 3CB) groups. With the same cutoff, no upregulated gene was identified in the sCR sample. With a more permissive threshold (fold change ≥1.5 or ≤ −1.5, nominal P < .001), 18 DEGs were identified. Among these 18 DEGs, 8 genes were upregulated in the CR CD138− baseline samples (supplemental Figure 3C). Interestingly, manual inspection of the upregulated DEGs in the CR baseline sample against currently available knowledge bases suggested that the upregulated DEGs play a role in immune regulation, such as TNFRSF13B and GNLY. GAGE12D, the most significantly upregulated gene in the sCR sample, belongs to the GAGE cancer/testis antigens gene family. GAGE proteins can elicit immune response and serve as potential targets for cancer immunotherapy.17 Gene set enrichment analysis showed a significant enrichment of genes in the MYC hallmark pathway (FDR q value = 0, NES = 3.4) and cell cycle–related pathway (HALLMARK_E2F_TARGETS, FDR q value = 0, NES = 2.6) (supplemental Figure 3D).
To further investigate changes in the tumor microenvironment, we performed transcriptome deconvolution analysis to infer the cellular composition of immune cells. A monocyte chemoattractant protein counter18 was used to produce the absolute abundance scores for 8 major immune cell types, endothelial cells, and fibroblasts (supplemental Figures 4A and 5A). Unsupervised clustering of the abundance scores grouped the specimens into 2 major clusters with distinct immune signatures. The abundance scores produced by the monocyte chemoattractant protein counter indicated that the abundance of cytotoxic lymphocytes, CD8 T cells, NK cells, myeloid dendritic cells (a major stimulator of T cells), monocytes linage, endothelial cells as well as the normal peritoneal fibroblasts, which were significantly lower in the CD138+ samples as expected. No significant association was observed between clinical response and cellular composition of tumor microenvironment even when pre- and posttreatment samples were arranged together (supplemental Figure 5A-B). In agreement with this, unsupervised clustering of immune genes clearly separated CD138+ samples from CD138− samples (supplemental Figures 4B and 5B).
Interferon-γ (IFN-γ) signature analysis
IFN-γ signature was previously reported to predict clinical response to PD-1 blockade in melanoma patients at baseline.19 We extracted the IFN-γ signature genes from the RNA-seq data and correlated gene signature expression profile at baseline with the level of change of tumor burden (M-protein with patients expressing a monoclonal protein and light chain for patients with light chain only disease) in those samples (Figure 2G). Spearman correlations between gene expression level and M protein level ≥0.3 or ≤ −0.3 were shown. We found the expression level of CXCR6, CD274 (PD-L1), PDCD1LG2, CD3D, CD27, and TAGAP were associated with increasing tumor burden and lack of response to treatment with pembrolizumab, whereas the expression level of CD2, GZMA, IL2RG, GZMB, TIGIT, HLA-DRB1, CMKLR1, and CD8B were associated with decreasing tumor burden and response to pembrolizumab.
Discussion
In our pilot study, only 1 patient (8%) achieved a response to single-agent pembrolizumab. It is remarkable, however, that this patient had a stringent complete response (CR) with NGS negativity at a level of 10−4 that is ongoing 27 months since treatment start date after only 3 cycles of single-agent pembrolizumab and without toxicities except mild laboratory abnormalities (Figure 2A-B). This patient experienced grade 2 transaminitis after 3 cycles, which resolved by stopping the study drug and did not require steroid therapy (supplemental Table 3). This patient had amplification of CKS1B, deletion 17p, and high-risk GEP70 score, all of which have been associated with worse clinical outcomes and resistance to therapy in newly diagnosed myeloma (NDMM) as well as decreased survival in smoldering myeloma and greater risk of progression to MM from SMM.20-22
Immune profiling of bone marrow samples by multiparametric flow cytometry showed that the extraordinary responder (patient 3) had high levels of PD-1 expression and low levels of other inhibitory receptors, such as TIM-3, LAG-3, and BTLA on CD8+ T cells compared with nonresponders at baseline (Figure 2E). Interestingly, the expression of various stimulatory and inhibitory receptors, such as CD137 (4-1BB), PD-1, TIM-3, CTLA-4, LAG-3, BTLA, and ICOS, increased markedly on CD8+ T cells in the bone marrow after therapy(Figure 2F). Whole transcriptome sequencing of bone marrow samples showed increased IFN-γ signature in the responder at baseline, a signature previously associated with response to pembrolizumab in solid tumors (Figure 2G).19 After pembrolizumab treatment, there was strong activation of immune stimulatory genes and decreased immune checkpoint genes in the responder compared with most nonresponders (Figure 2H). Taken together, these data suggest that the extraordinary responder had a preexisting immune response in the bone marrow that was enhanced with pembrolizumab therapy, which is consistent with the known mechanism of action of this agent. In contrast, nonresponders had higher levels of multiple immune checkpoint genes in the bone marrow compared with the responder, suggesting their T cells were more exhausted and may require combination approaches targeting multiple immune checkpoints to restore function.23
The responder also had a distinct GEP70 profile consistent with high-risk disease (supplemental Figure 2). Deconvolution of the bone marrow transcriptome before and after treatment did not reveal any association between microenvironment composition and clinical outcome (supplemental Figures 4 and 5). The most upregulated DEGs in tumor cells in the patient who progressed to myeloma were FGFR3, WHSC1, and MAF, which were reported to be associated with t4,14 translocation in MM.16 In the responder, the immune genes TNFRSF13B and GNLY were upregulated besides GAFE12D, which encodes a cancer/testis antigen (supplemental Figure 3). WES revealed the baseline mutational profile of the patients with high rates of BRAF (27%), FOXO3 (27%), and KRAS (27%) mutations, which have been described before in myeloma24,25; however, mutational load did not correlate with response (Figure 3). For example, patients 6 and 7 progressed either while on therapy or shortly after therapy had been discontinued and had higher mutational load than patient 3, who achieved an sCR. Neoantigen analysis prediction in patients with NDMM showed that the average neoantigen load was 23.5226 and that patients with greater than average load had worse PFS. To our knowledge, neoantigen analysis of SMM has not been reported yet. In this study, the average neoantigen load was 11.27 for strong-binder and 9.45 for weak-binder, and neither correlated with response to pembrolizumab (Figure 3).
To our knowledge, this is the first time that an sCR is reported with pembrolizumab monotherapy and without radiation in myeloma. Previously, a patient with relapsed/refractory myeloma achieved initially SD with nivolumab (anti–PD-1 monoclonal antibody) monotherapy. The patient then had an increase in serum free light chain ratio and increasingly symptomatic rib plasmacytoma and was treated with radiation therapy to the plasmacytoma with resolution of symptoms. He was assessed in the study as being in CR on the basis of the rib lesion having become asymptomatic.27 In our report, sCR was reached in a patient who not only was at high risk of progression to multiple but also had genetic aberrancies that meet the definition of high-risk MM (high score GEP70 and deletion 17p on FISH). The sCR was achieved without radiation or other therapies, which could have confounded the CR. Similarly to the other report of CR in anti–PD-1 monotherapy in myeloma, these were extraordinary responders, and no other patients achieved deep responses with single-agent anti–PD-1 monoclonal antibody treatment.
In 2 randomized phase 3 studies in relapsed/refractory myeloma and NDMM, pomalidomide/dexamethasone and lenalidomide/dexamethasone, respectively, with or without pembrolizumab led to worse PFS and overall survival in the pembrolizumab arms.10 Because of this, our study was placed on hold, and 13 of the 16 planned patients were treated. As of the date of publication of this article, the authors have not requested that the FDA lift the hold to continue enrollment. This is based on the low efficacy with only 1 responder out of 13. The FDA has signaled that development of immune therapies in myeloma can continue with adequate patient selection and safety. We expect that newer immunotherapeutic strategies will be developed in myeloma with the guidance of the FDA and other regulatory authorities.10
Strategies using other immunotherapeutics, such as vaccines (PVX-410), elotuzumab, and daratumumab, have reported ORR of 0%, 10%, and 56% and CR rate of 0%, 0% and 5%, respectively.28-30 Emerging data from the use of myeloma-like regimens, such as carfilzomib, lenalidomide, and dexamethasone with or without autologous stem cell transplantation, have reported high ORR of 98% and 100%, and MRD-negative CR of 55% and 63%, respectively.31,32 However, it is unclear at this time whether the morbidity associated with such intensive therapies is justified for SMM. Despite the small sample size of our study, the sCR observed in 1 patient together with the immune and genomic profiling analyses suggests that SMM may be naturally immunogenic in a small subset of patients, and immunotherapy strategies aimed at enhancing antitumor T-cell responses need to be explored for immunoprevention of SMM. Toward this goal, we have initiated a clinical trial to evaluate neoantigen vaccines in SMM (#NCT03631043).
Supplementary Material
The full-text version of this article contains a data supplement.
Acknowledgments
The authors thank the participating patients and their families.
This work was supported in part by The Anderson Cancer Center Support Grant (National Institutes of Health, National Cancer Institute P30 CA016672), the Leukemia and Lymphoma Society Specialized Center of Research (LLS SCOR), the Dr Miriam and Sheldon G. Adelson Medical Research Foundation, the Multiple Myeloma Research Foundation, and The University of Texas Anderson Moon Shot Program.
The trial collaborator (Merck) provided pembrolizumab and partial financial support but was not involved in study design, data collection, analysis, interpretation, writing of this report, or decision to submit the manuscript for publication. The trial collaborator reviewed this report for accuracy. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Authorship
Contribution: E.E.M. designed the research, treated patients, interpreted the data, and wrote the manuscript; H.L., D.M.W., and B.A. treated patients, interpreted the data, and provided manuscript comments; L.F. analyzed the data; Z.B. interpreted the data and wrote the manuscript; G.H., R.M., Y.Q., Z.Z., K.E., S.Z., J.Z., X.S., X.M., M.M., V.B., A.F., and L.W. did correlative studies, interpreted the data, and provided manuscript comments; S.S.N. designed the research, interpreted the data, and wrote the manuscript; and R.Z.O. designed the research, treated patients, interpreted the data, and provided manuscript comments.
Conflict-of-interest disclosure: E.E.M. has received research support from Sanofi, Quest Diagnostics, Novartis, JW Pharma, and Merck and consultant fees from Takeda, Celgene, Sanofi, Seattle Genetics, and BMS. R.M. is an employee at Celularity. H.L. has received consulting fees from Adaptive Biotechnologies, Celgene, Pimera, and Takeda and research support from Amgen, Daiichi Sankyo, Janssen, and Takeda. S.S.N. has received research support from Kite/Gilead, Celgene, Cellectis, Poseida, Merck, Acerta, Karus, and BMS and served as consultant and advisory board member for Kite/Gilead, Celgene, Novartis, Unum Therapeutics, Pfizer, CellMedica, and Merck. R.Z.O. has received consulting fees from Amgen, Bristol-Myers-Squibb, Celgene, Janssen, Kite Pharma, Sanofi, and Takeda and research support from Amgen, BioTheryX, and Spectrum Pharmaceuticals. The remaining authors declare no competing financial interests.
Correspondence: Elisabet E. Manasanch, Department of Lymphoma/Myeloma, The University of Texas MD Anderson Cancer Center, 1515 Holcombe Blvd, Unit 429, Houston, TX 77030; e-mail: eemanasanch@mdanderson.org.
References
- 1.Dhodapkar MV, Sexton R, Das R, et al. . Prospective analysis of antigen-specific immunity, stem-cell antigens, and immune checkpoints in monoclonal gammopathy. Blood. 2015;126(22):2475-2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dhodapkar MV, Sexton R, Waheed S, et al. . Clinical, genomic, and imaging predictors of myeloma progression from asymptomatic monoclonal gammopathies (SWOG S0120). Blood. 2014;123(1):78-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jelinek T, Paiva B, Hajek R. Update on PD-1/PD-L1 inhibitors in multiple myeloma. Front Immunol. 2018;9:2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu J, Hamrouni A, Wolowiec D, et al. . Plasma cells from multiple myeloma patients express B7-H1 (PD-L1) and increase expression after stimulation with IFN-γ and TLR ligands via a MyD88-, TRAF6-, and MEK-dependent pathway. Blood. 2007;110(1):296-304. [DOI] [PubMed] [Google Scholar]
- 5.Paiva B, Azpilikueta A, Puig N, et al. . PD-L1/PD-1 presence in the tumor microenvironment and activity of PD-1 blockade in multiple myeloma. Leukemia. 2015;29(10):2110-2113. [DOI] [PubMed] [Google Scholar]
- 6.Kelly KR, Espitia CM, Zhao W, et al. . Oncolytic reovirus sensitizes multiple myeloma cells to anti-PD-L1 therapy. Leukemia. 2018;32(1):230-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Merck Keytruda package insert 2018. Available at: https://www.merck.com/product/usa/pi_circulars/k/keytruda/keytruda_pi.pdf.
- 8.Kumar S, Paiva B, Anderson KC, et al. . International Myeloma Working Group consensus criteria for response and minimal residual disease assessment in multiple myeloma. Lancet Oncol. 2016;17(8):e328-e346. [DOI] [PubMed] [Google Scholar]
- 9.FDA Alerts Healthcare Professionals and Oncology Clinical Investigators about Two Clinical Trials on Hold Evaluating KEYTRUDA® (pembrolizumab) in Patients with Multiple Myeloma 2017. Available at: https://www.fda.gov/drugs/drug-safety-and-availability/fda-alerts-healthcare-professionals-and-oncology-clinical-investigators-about-two-clinical-trials.
- 10.Gormley NJ, Pazdur R. Immunotherapy combinations in multiple myeloma - known unknowns. N Engl J Med. 2018;379(19):1791-1795. [DOI] [PubMed] [Google Scholar]
- 11.Yang Z, Jiang X, Jiang X, Zhao H. X-inactive-specific transcript: a long noncoding RNA with complex roles in human cancers. Gene. 2018;679:28-35. [DOI] [PubMed] [Google Scholar]
- 12.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. ; ICGC PedBrain . Signatures of mutational processes in human cancer [published correction appears in Nature. 2013;502(7470):258]. Nature. 2013;500(7463):415-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alexandrov LB, Jones PH, Wedge DC, et al. . Clock-like mutational processes in human somatic cells. Nat Genet. 2015;47(12):1402-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bolli N, Maura F, Minvielle S, et al. . Genomic patterns of progression in smoldering multiple myeloma. Nat Commun. 2018;9(1):3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Auslander N, Zhang G, Lee JS, et al. . Robust prediction of response to immune checkpoint blockade therapy in metastatic melanoma [published correction appears in Nat Med. 2018;24(12):1942]. Nat Med. 2018;24(10):1545-1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalff A, Spencer A. The t(4;14) translocation and FGFR3 overexpression in multiple myeloma: prognostic implications and current clinical strategies. Blood Cancer J. 2012;2(9):e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gjerstorff MF, Ditzel HJ. An overview of the GAGE cancer/testis antigen family with the inclusion of newly identified members. Tissue Antigens. 2008;71(3):187-192. [DOI] [PubMed] [Google Scholar]
- 18.Becht E, Giraldo NA, Lacroix L, et al. . Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression [published correction appears in Genome Biol. 2016;17(1):249]. Genome Biol. 2016;17(1):218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ayers M, Lunceford J, Nebozhyn M, et al. . IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest. 2017;127(8):2930-2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Claussen CM, Lee H, Shah JJ, et al. . Gene expression profiling predicts clinical outcomes in newly diagnosed multiple myeloma patients in a standard of care setting [abstract] Blood. 2016;128(22). Abstract 5628. [Google Scholar]
- 21.Sonneveld P, Avet-Loiseau H, Lonial S, et al. . Treatment of multiple myeloma with high-risk cytogenetics: a consensus of the International Myeloma Working Group. Blood. 2016;127(24):2955-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dispenzieri A, Stewart AK, Chanan-Khan A, et al. . Smoldering multiple myeloma requiring treatment: time for a new definition? Blood. 2013;122(26):4172-4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dempke WCM, Fenchel K, Uciechowski P, Dale SP. Second- and third-generation drugs for immuno-oncology treatment—The more the better? Eur J Cancer. 2017;74:55-72. [DOI] [PubMed] [Google Scholar]
- 24.Lohr JG, Stojanov P, Carter SL, et al. ; Multiple Myeloma Research Consortium . Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer Cell. 2014;25(1):91-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walker BA, Wardell CP, Murison A, et al. . APOBEC family mutational signatures are associated with poor prognosis translocations in multiple myeloma. Nat Commun. 2015;6(1):6997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller A, Asmann Y, Cattaneo L, et al. ; MMRF CoMMpass Network . High somatic mutation and neoantigen burden are correlated with decreased progression-free survival in multiple myeloma. Blood Cancer J. 2017;7(9):e612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lesokhin AM, Ansell SM, Armand P, et al. . Nivolumab in patients with relapsed or refractory hematologic malignancy: preliminary results of a phase Ib study. J Clin Oncol. 2016;34(23):2698-2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jagannath S, Laubach J, Wong E, et al. . Elotuzumab monotherapy in patients with smouldering multiple myeloma: a phase 2 study. Br J Haematol. 2018;182(4):495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Landgren O, Cavo M, Chari A, et al. Updated results from the phase 2 centaurus study of daratumumab (DARA) monotherapy in patients with intermediate-risk or high-risk smoldering multiple myeloma (SMM) [abstract]. Blood 2018;132(suppl 1). Abstract 1994.
- 30.Nooka AK, Wang ML, Yee AJ, et al. . Assessment of safety and immunogenicity of PVX-410 vaccine with or without lenalidomide in patients with smoldering multiple myeloma: a nonrandomized clinical trial. JAMA Oncol. 2018;4(12):e183267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mailankody S, Kazandjian D, Korde N, et al. . Baseline mutational patterns and sustained MRD negativity in patients with high-risk smoldering myeloma. Blood Adv. 2017;1(22):1911-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mateos MV, Martinez-Lopez J, Rodriguez Otero P, et al. . Curative strategy (GEM-CESAR) for high-risk smoldering myeloma (SMM): carfilzomib, lenalidomide and dexamethasone (KRd) as induction followed by HDT-ASCT, consolidation with Krd and maintenance with Rd [abstract]. Blood. 2018. Abstract 731. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.