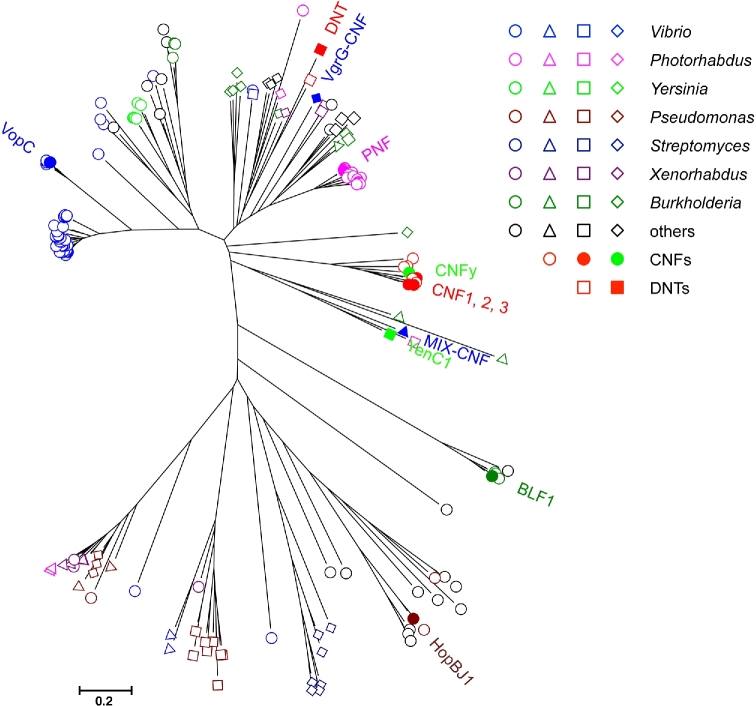
Figure 4.
Molecular phylogenetic relationships among the extended members of the CNF-domain-containing family. 167 representative sequences were collected from NCBI BLASTp search using VopC, DNT, CNF1, MIX-CNF, BLF1 and HopBJ1 as query, followed by screening for Gly-Cys-(Xaa)n-His motif, where n = 10–18, and trimming to within 150-residues flanking the motif. Sequences were aligned with MUSCLE (Edgar 2004), and then refined with Clustal W (Thompson, Higgins and Gibson 1994), before generating a Neighbor-Joining tree using MEGA7 (Kumar, Stecher and Tamura 2016). In some cases, Jalview (Waterhouse et al.2009) was used for editing the alignments. Those with known CNF1-like deamidase activity and a few select ones of interest (PNF, VgrG-CNF and YenC1) are indicated with solid symbols. Open red circles and squares are untested CNF or DNT sequences, respectively. Otherwise, open circles denote suspected T3SS effectors; triangles denoted suspected T6SS effectors; squares denote suspected large, multi-modular cytotoxins; and diamonds denote suspected Rhs-containing cytotoxins. Colors correspond to the indicated genera.