Figure 2.
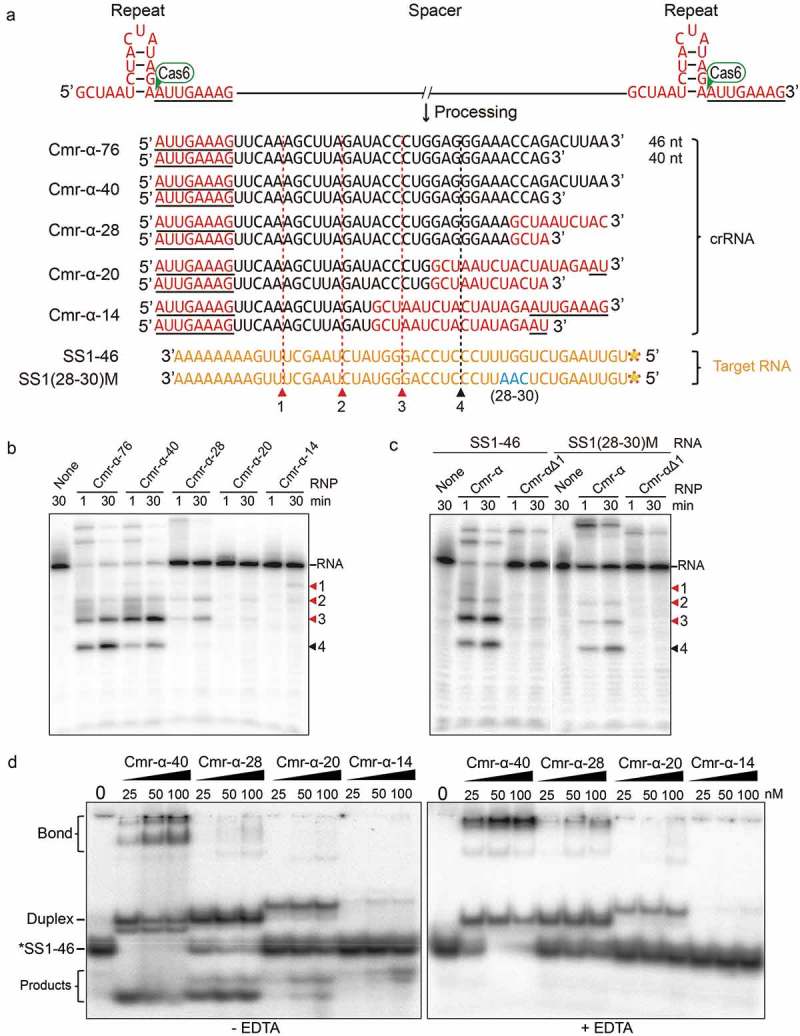
Influence of spacer length on in vitro RNA cleavage and RNA binding activity by the Cmr-α system.
(a) Predicted hairpin conformation of S. islandicus REY15A repeat elements (in red) and sequence of mature 40/46nt crRNAs with varying length of spacers. The primary processing site relying on Cas6 is indicated with a green triangle. Underlined bases indicate the 8 nt tag of the repeat sequence at the 5ʹ-end after primary processing. Two target RNAs used in the assay are shown in yellow. Predicted cleavage sites are indicated with red (for RNP complexes with 40/46 nt crRNAs) and black (only for RNP complexes with 46 nt crRNAs) triangles. (b) In vitro RNA cleavage assay using the Cmr-α complexes purified from each strain harbouring different lengths of spacers (76 nt, 40 nt, 28 nt, 20 nt and 14 nt). Fifty nanomolars of each Cmr-α complex was incubated with 25 nM labelled SS1-46 RNA for indicated time points. Then, the samples were analysed by denaturing PAGE. (c) Comparison of RNA cleavage on SS1-46 RNA and a trinucleotide mutation substrate (28–30 nt) by Cmr-α and Cmr-αΔ1 complexes. Cmr-αΔ1 – Cmr-α complex without Cmr1α, which was purified from S. islandicus ΔCmr1α. (d) Target RNA binding assay of Cmr-α complex with varying length of spacers (40 nt, 28 nt, 20 nt and 14 nt) for indicated amounts of SS1-46 RNA. Left: binding buffer without EDTA, Right: binding buffer with EDTA.
