Figure 1.
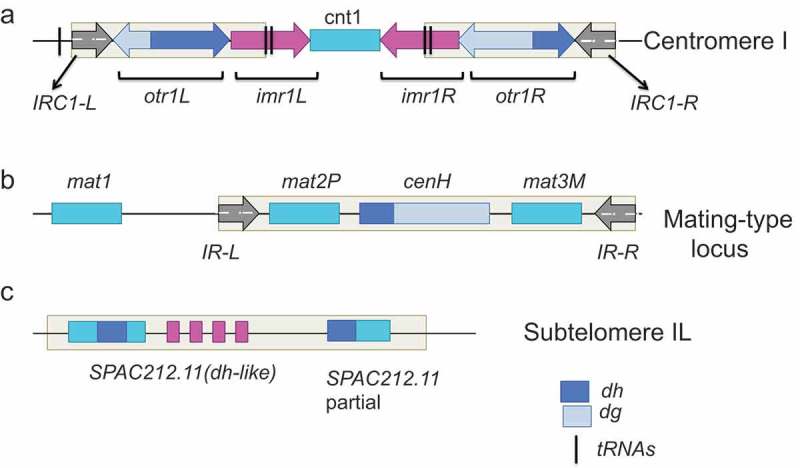
Sites of heterochromatin assembly in the fission yeast genome. This figure is adapted from [54] (a). Fission yeast centromeres contain a unique central domain consisting of central core region (cnt) and inner repeat sequences (imr), which is flanked by the outer repeat elements composed of one or more tandem array dg and dh repeats. Clusters of tRNA genes and/or IRC inverted repeats are present at the border of the pericentromeric heterochromatin. The number of dg/dh repeats differs between each chromosome arm, from 1 (cen1L) to 7–8 (cen3R) (b). At the mating type locus, mat2 and mat3 genes are located within a 20-kb heterochromatin domain. RNA interference regulates assembly of heterochromatin on the cenH element that shares strong homology with dg and dh centromeric repeats. Heterochromatin can also be nucleated in an RNA interference-independent manner within the mat2P/mat3M locus, including a 2.1 Kb region between cenH and mat3 by Atf1/Pcr1. The heterochromatic domain is restricted by boundary elements IR-L and IR- R. (c). cenH-like elements/dh-like sequences (SPAC212.11) within tlh1 and tlh2 genes are located at subtelomeric regions, which can nucleate heterochromatin both in an RNA interference-dependent and RNA interference-independent manner by the telomere specific factor Taz1.
